FULL TITLE:
A Multi-modal Parcellation of Human Cerebral Cortex
SPECIES:
Human
DESCRIPTION:
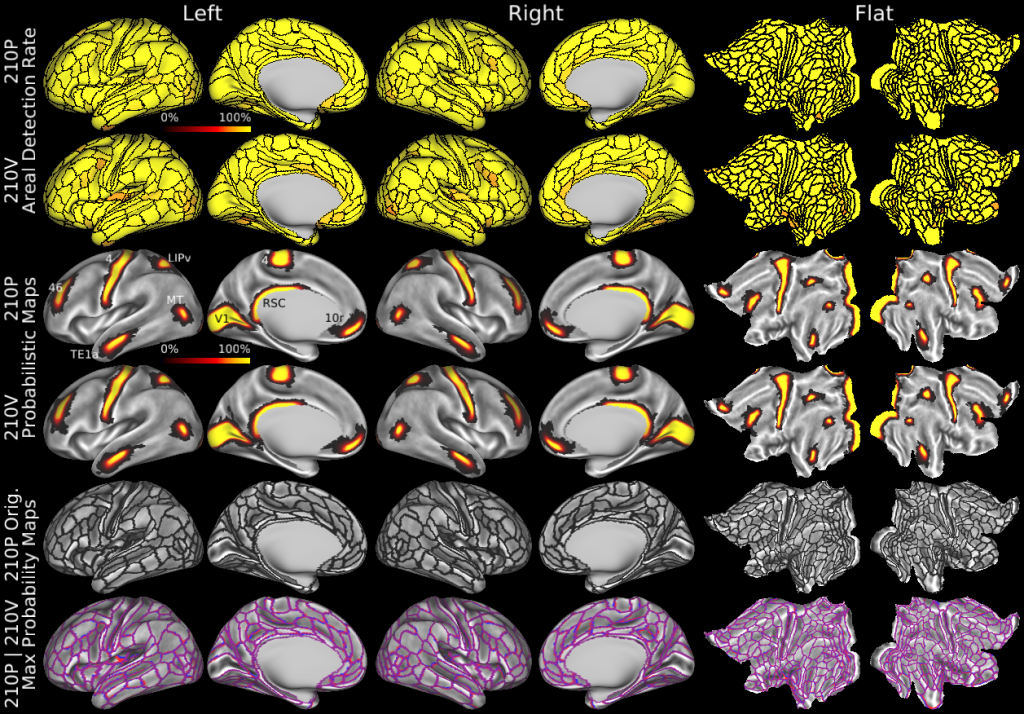
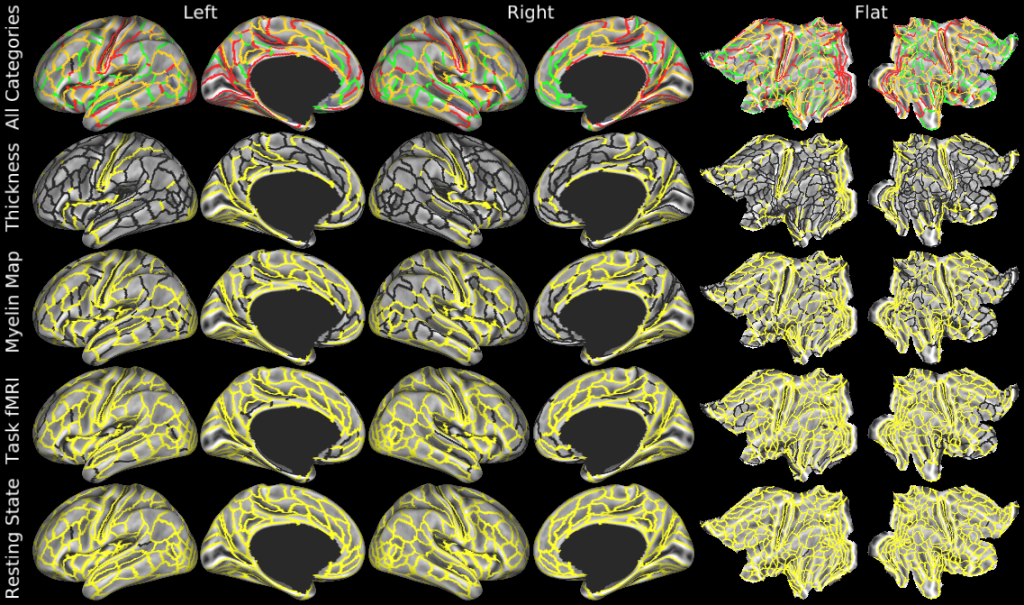
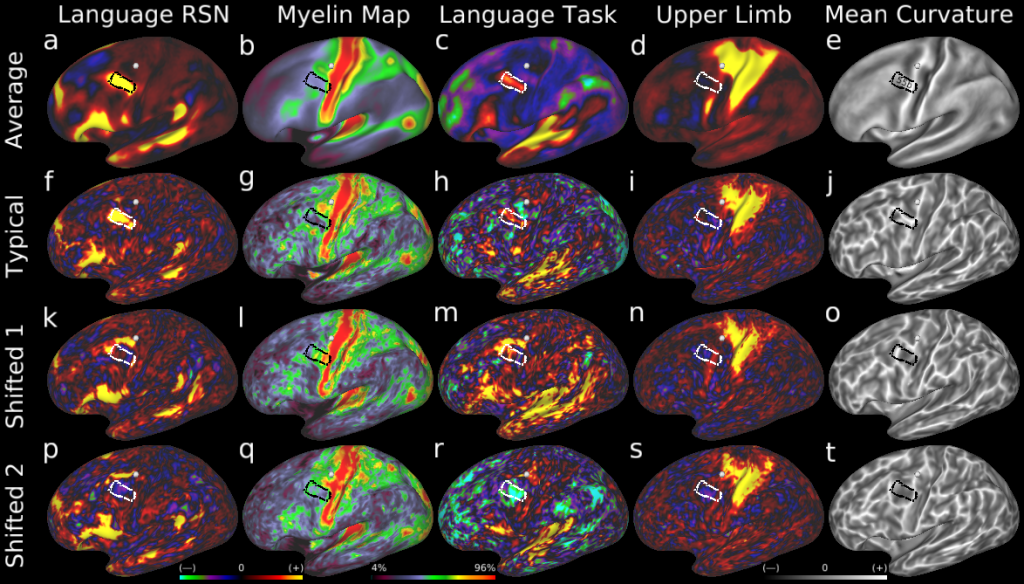
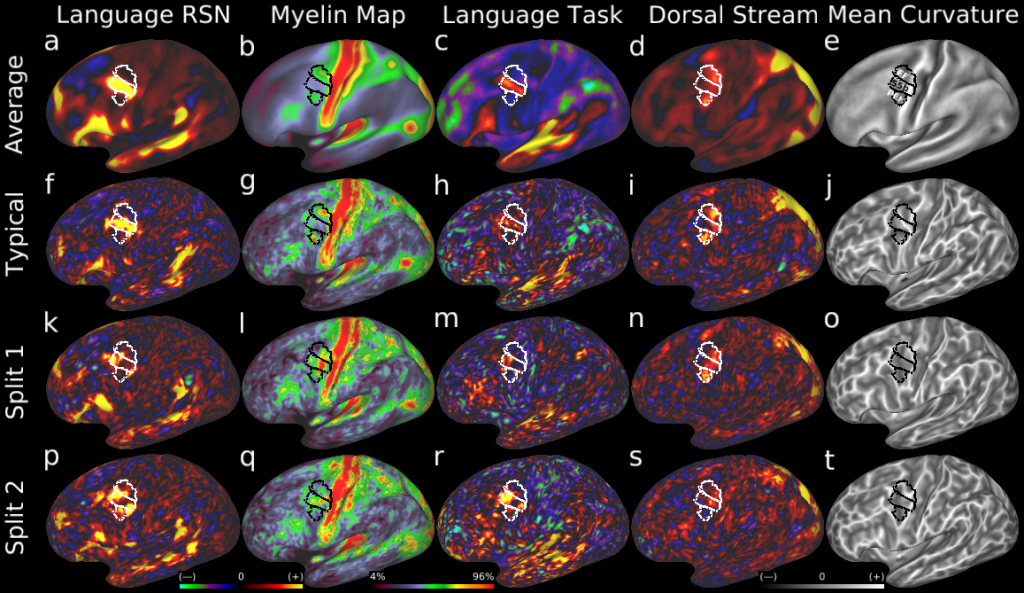
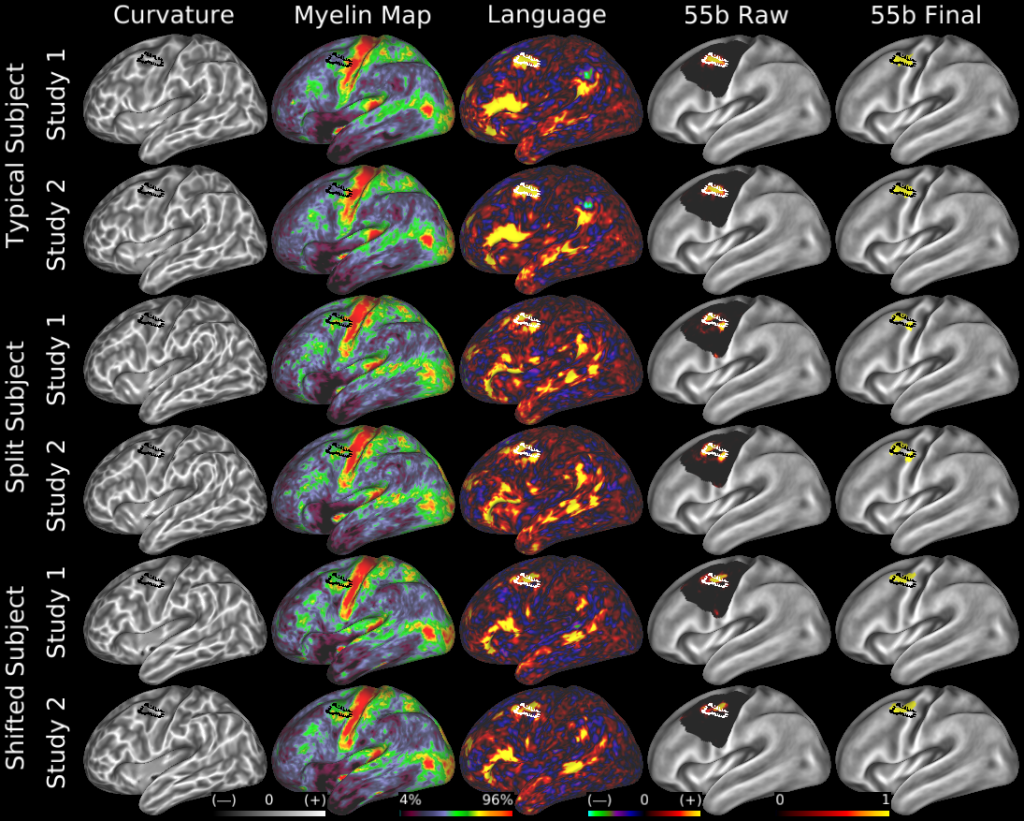
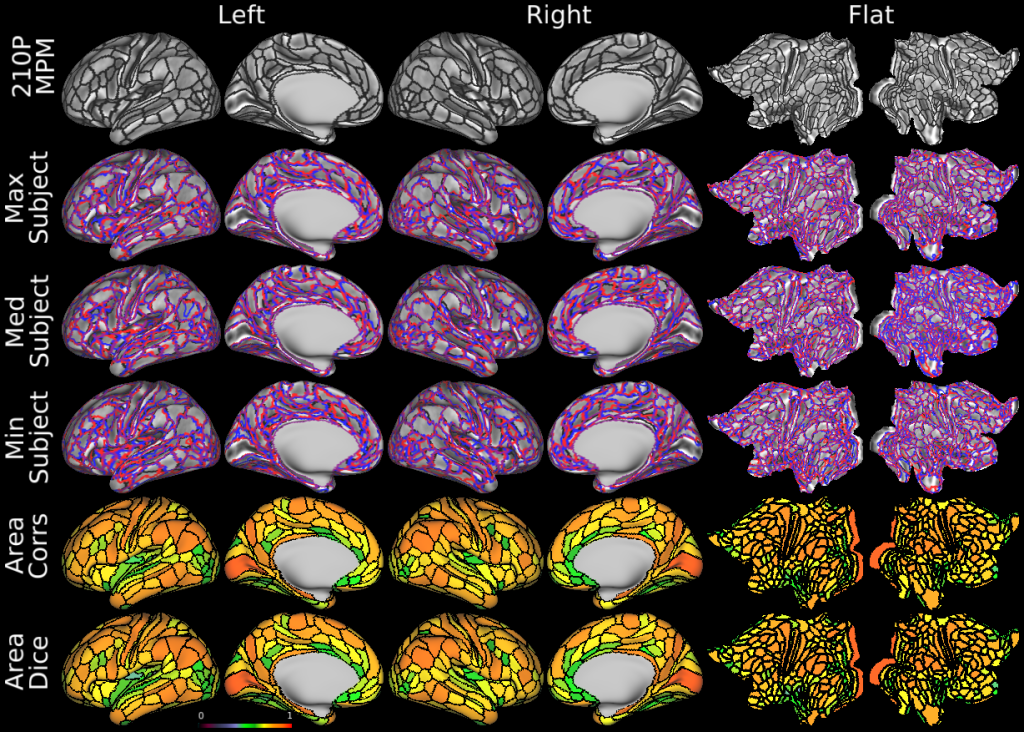
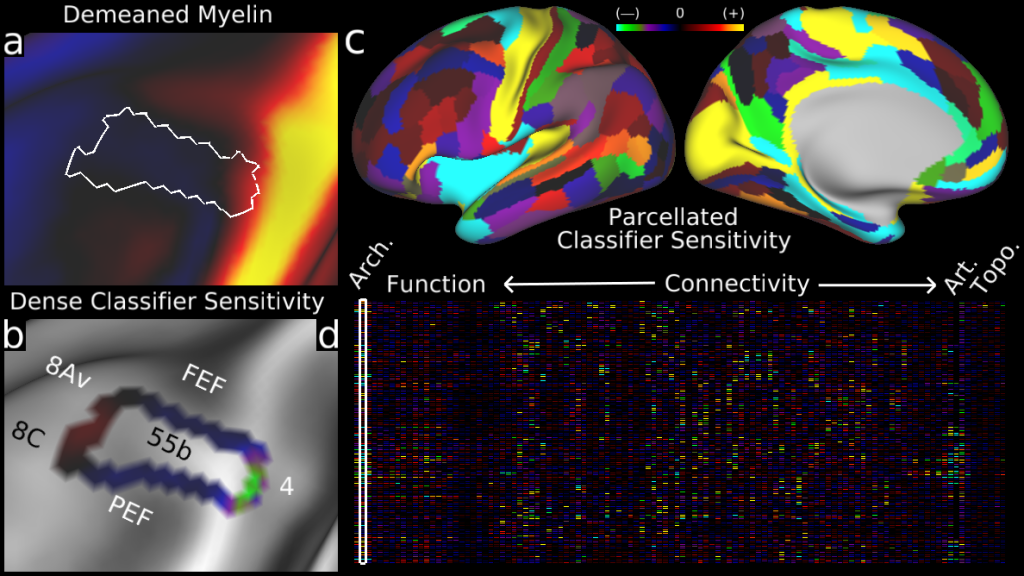
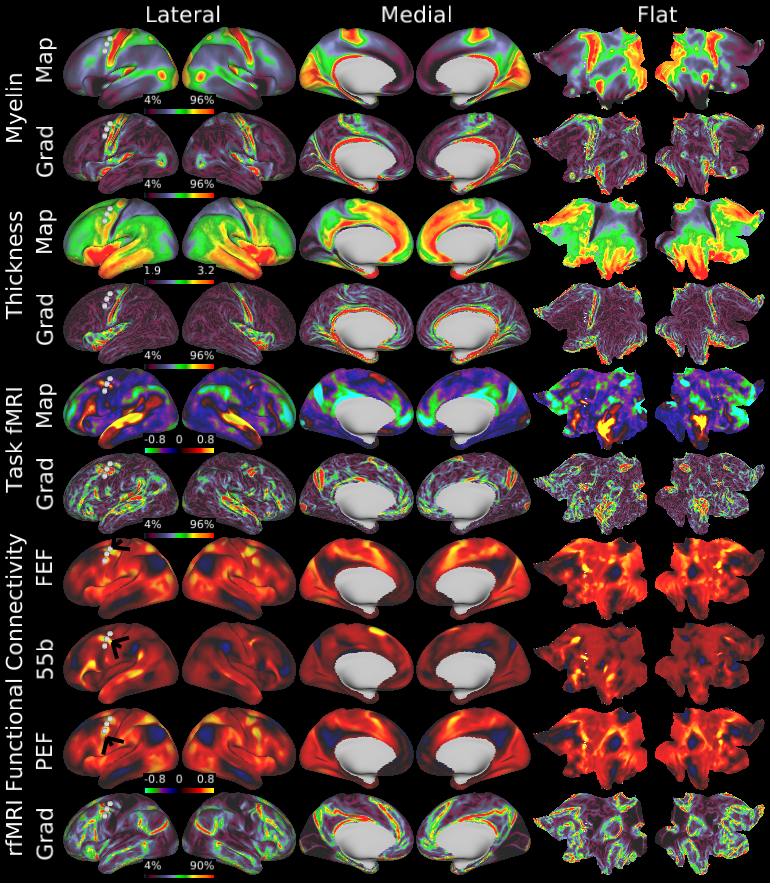
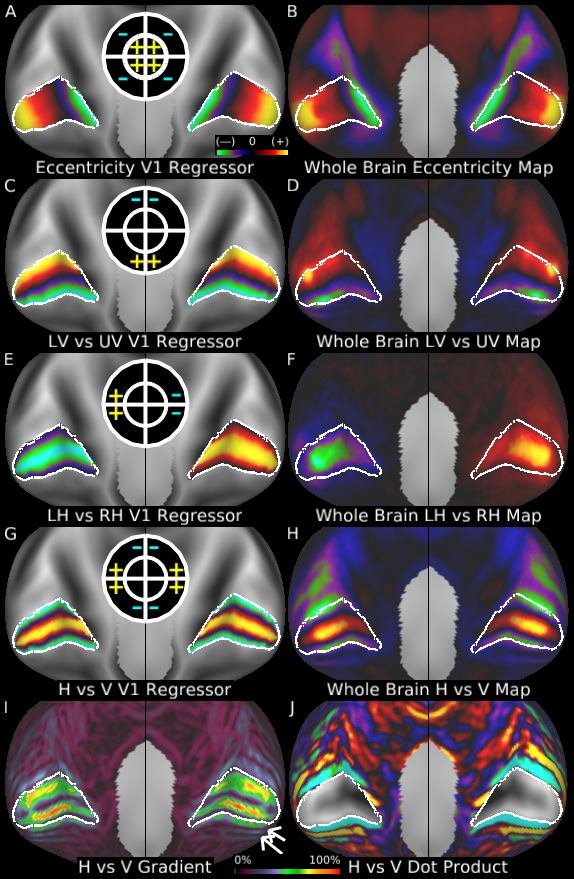
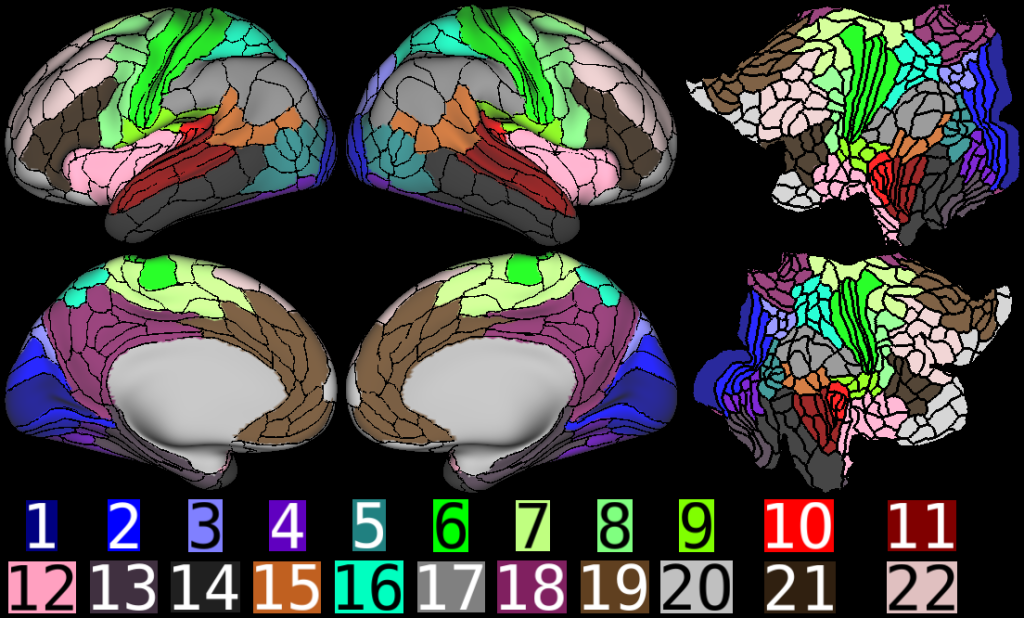
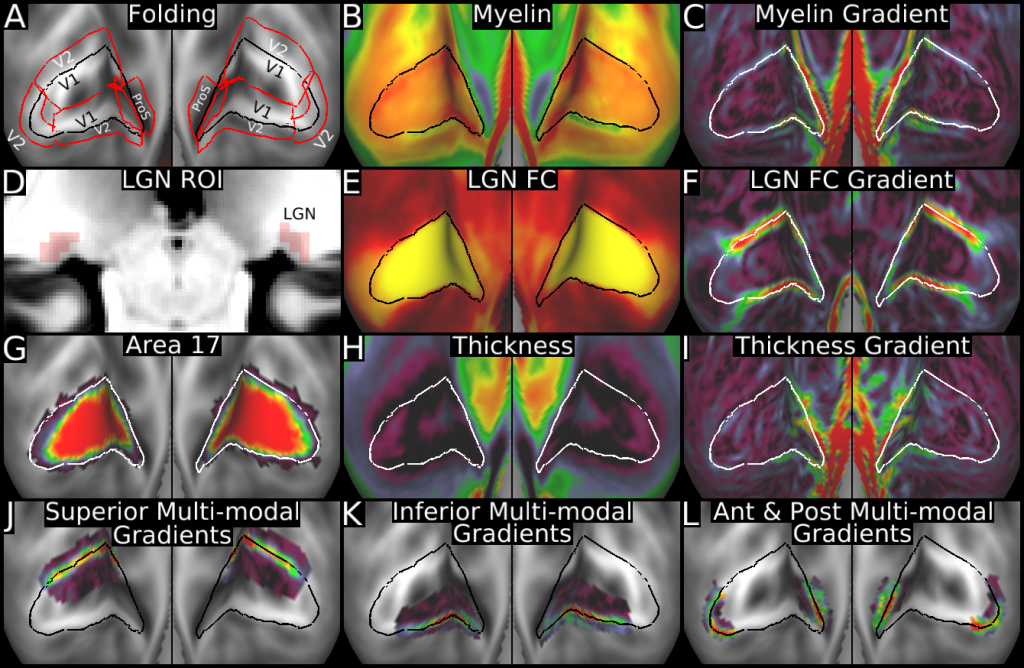
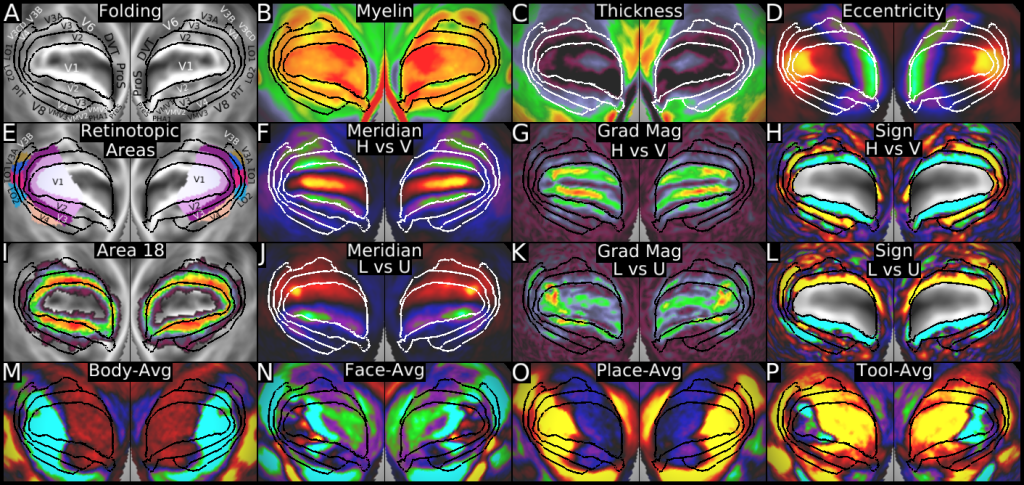
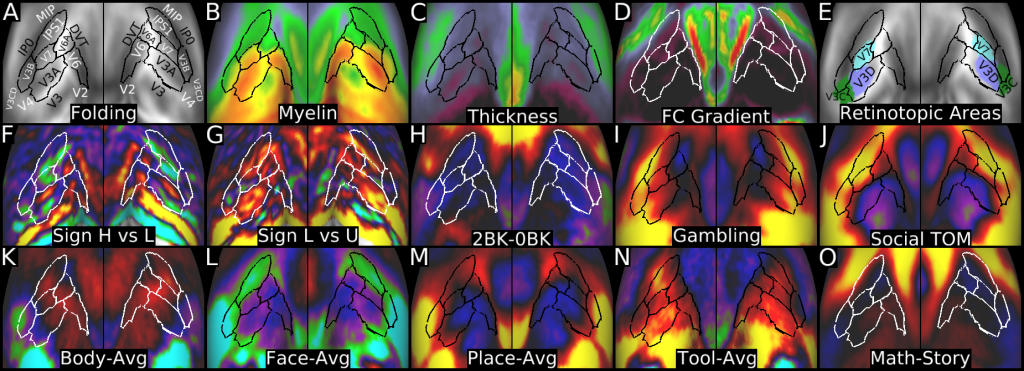
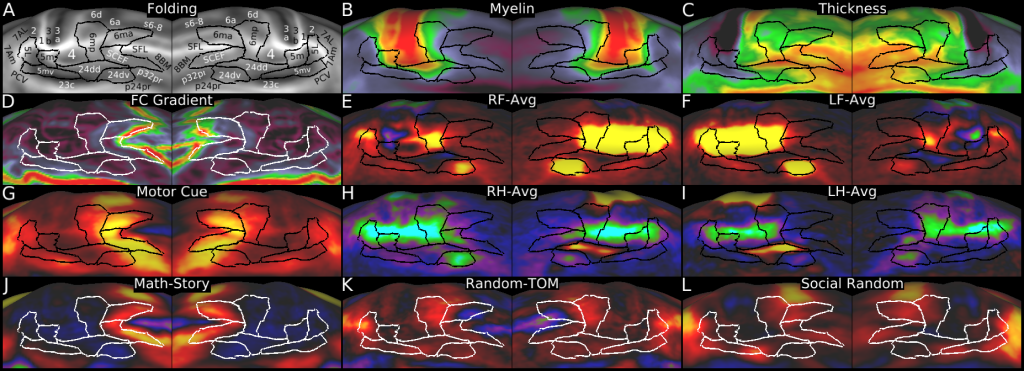
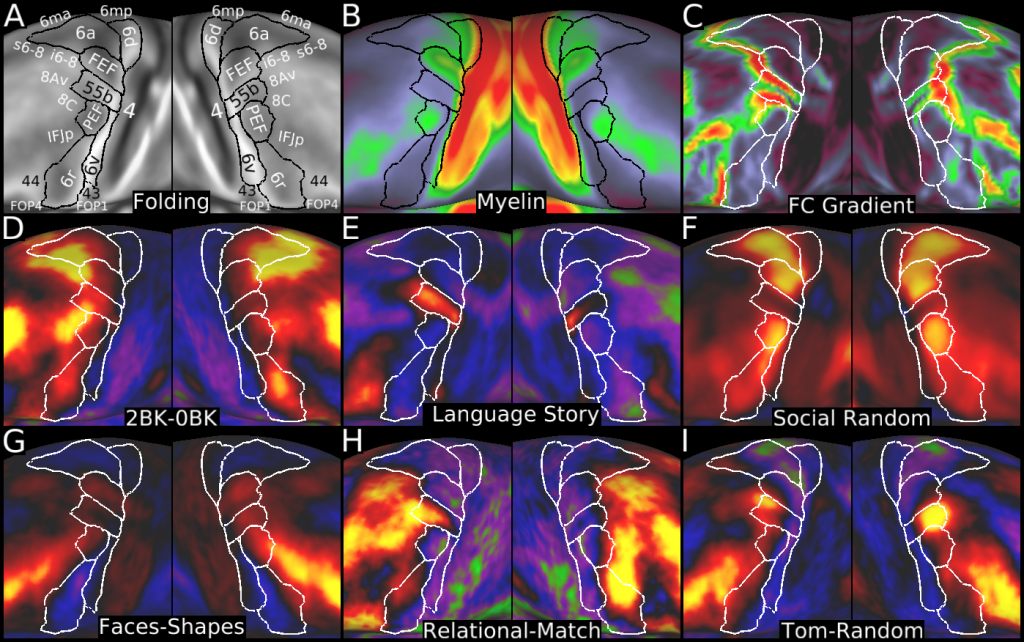
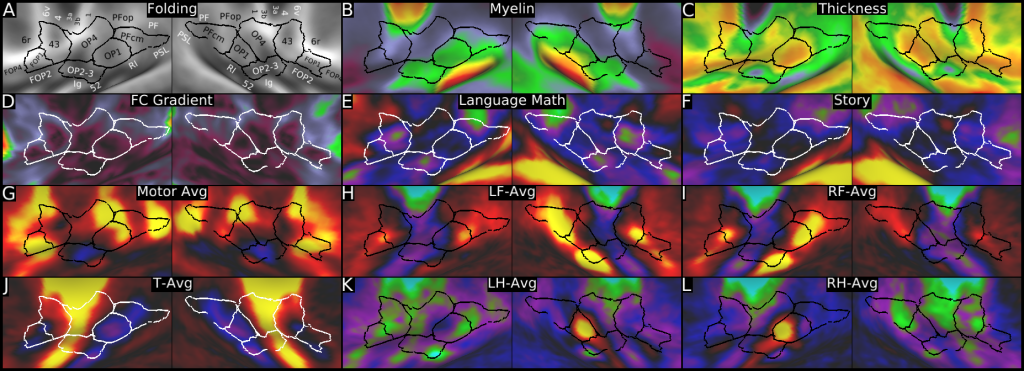
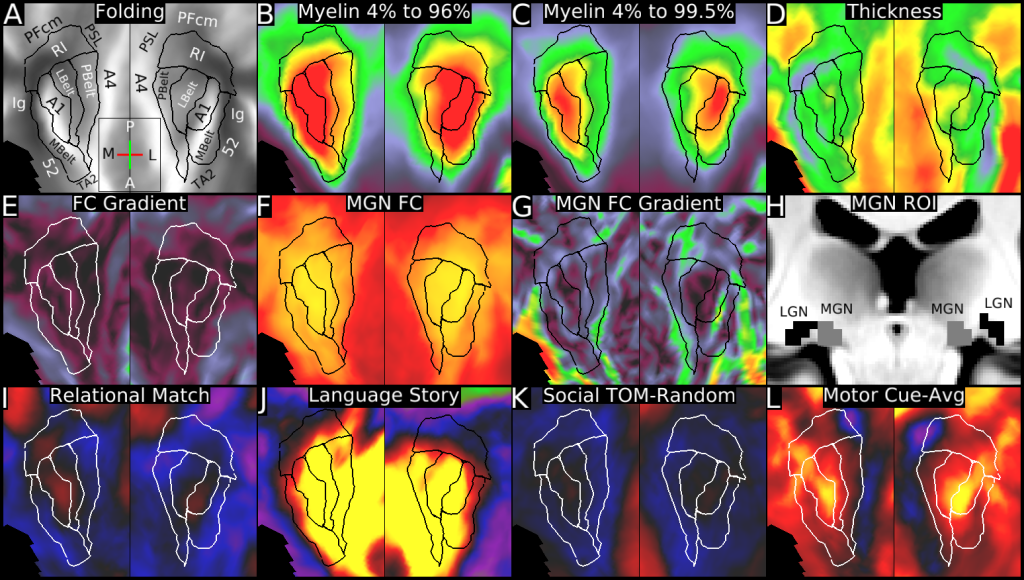
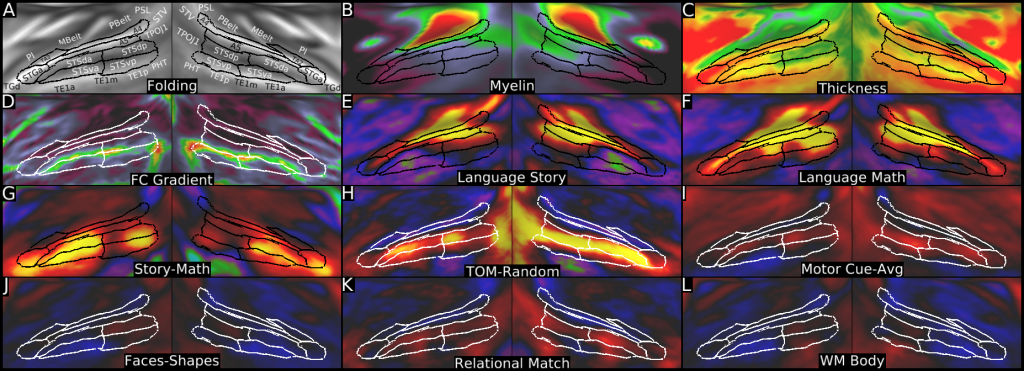
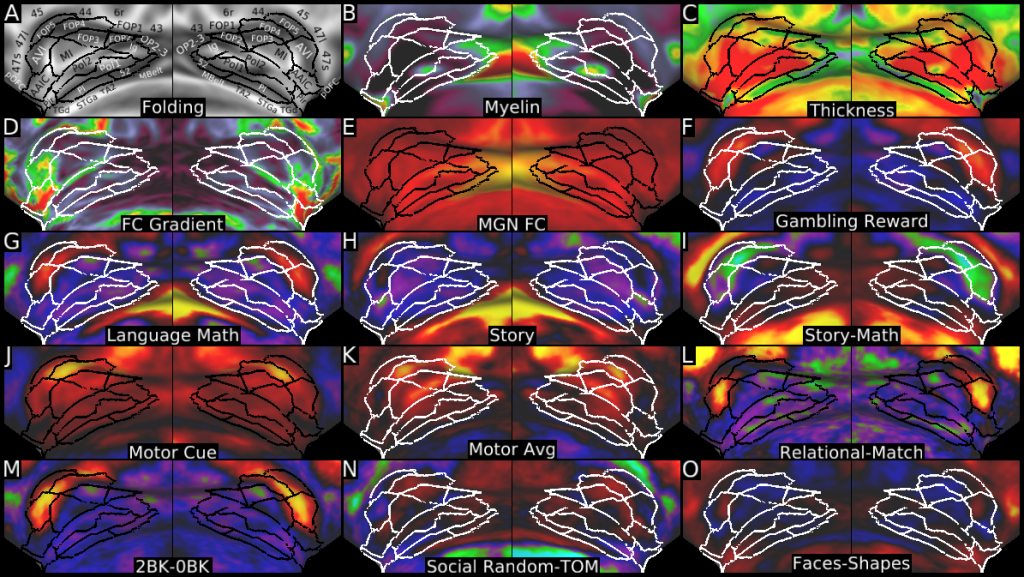
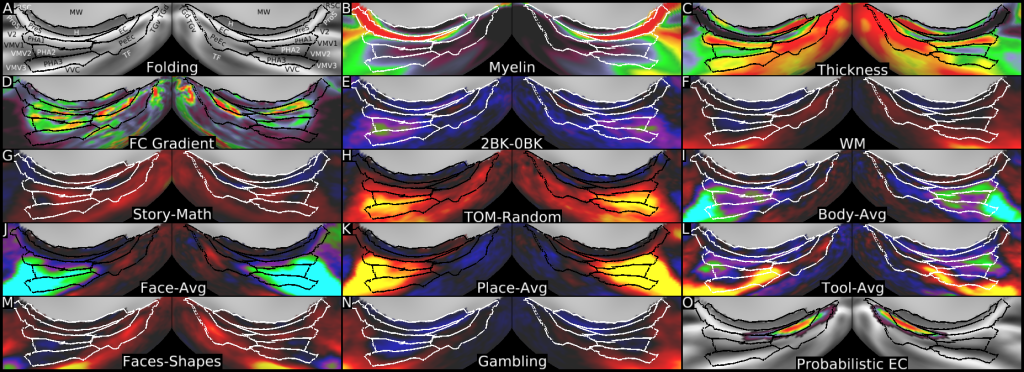
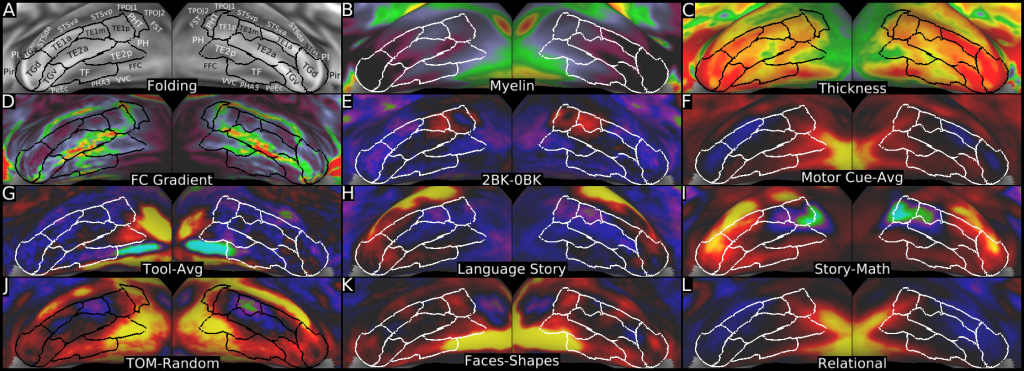
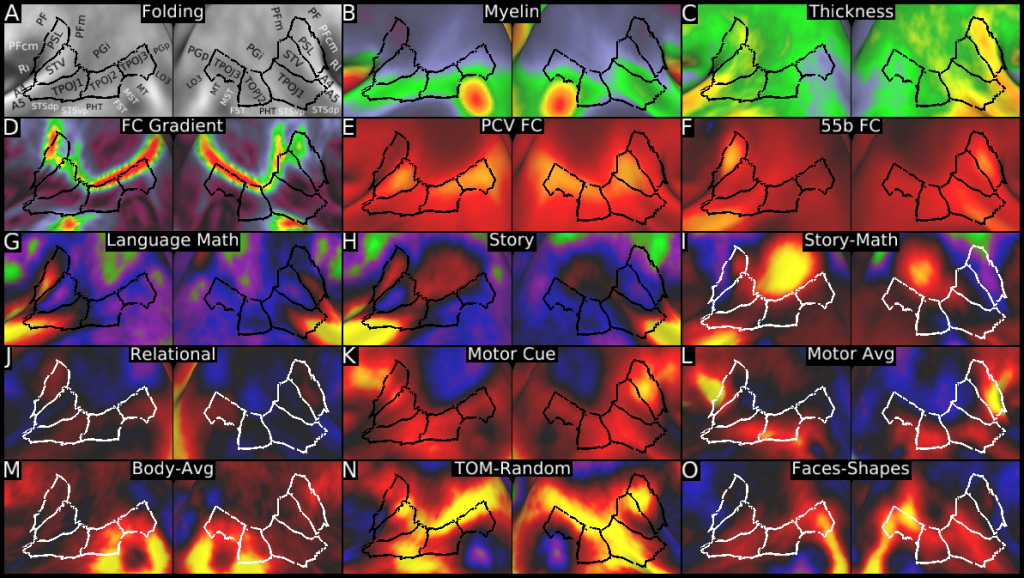
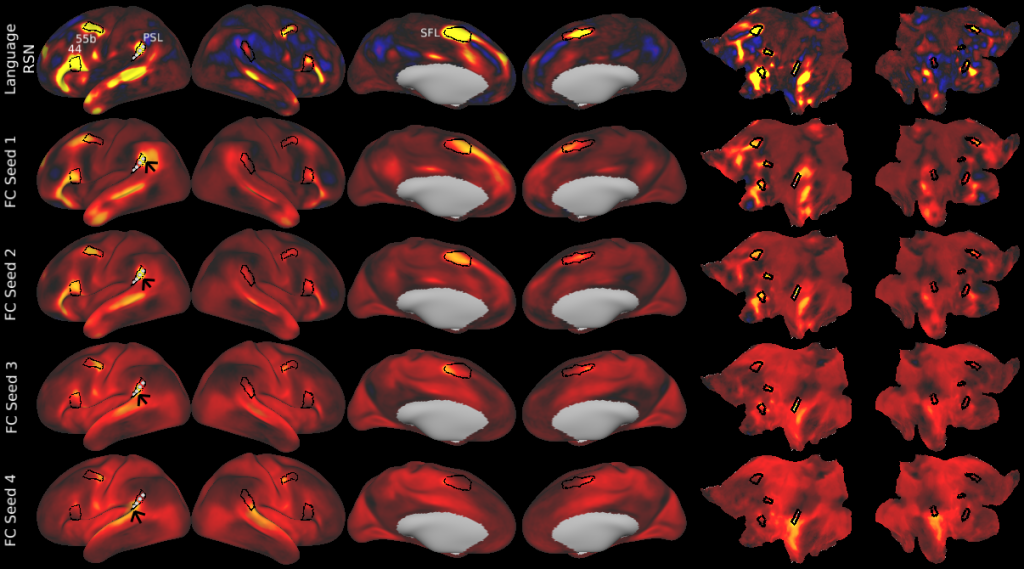
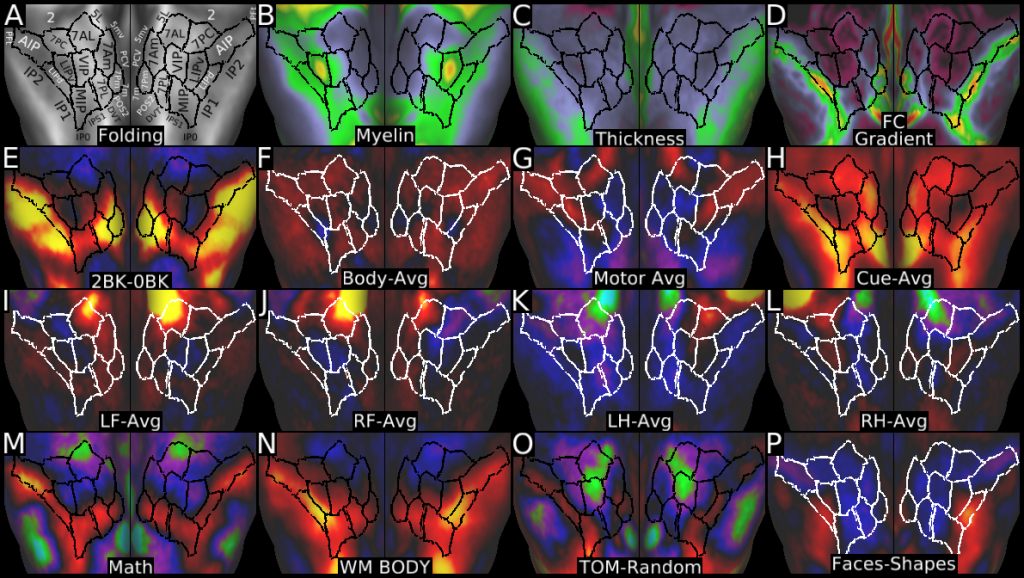
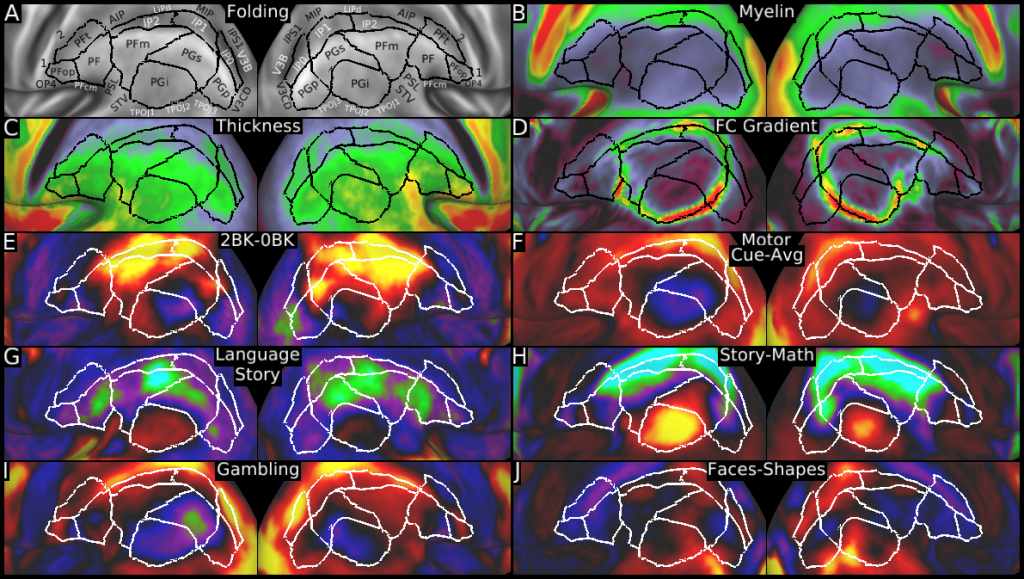
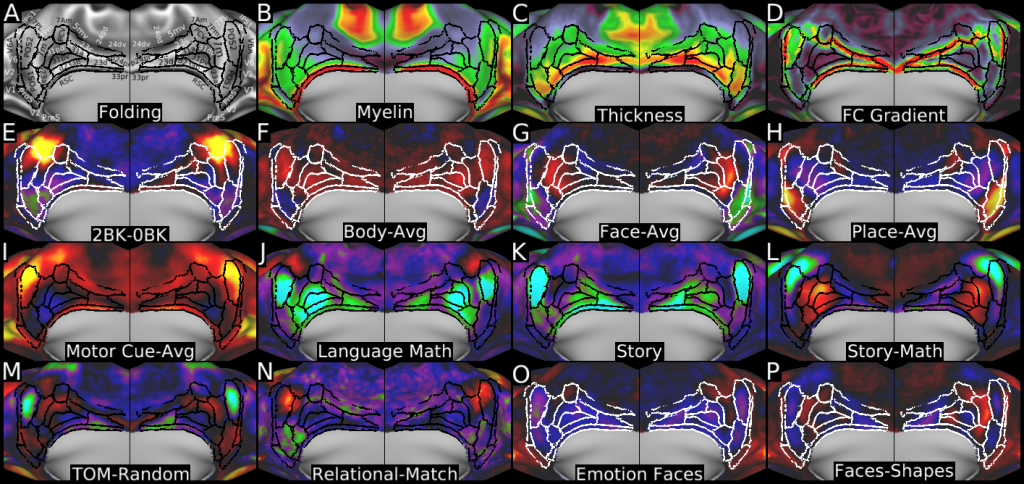
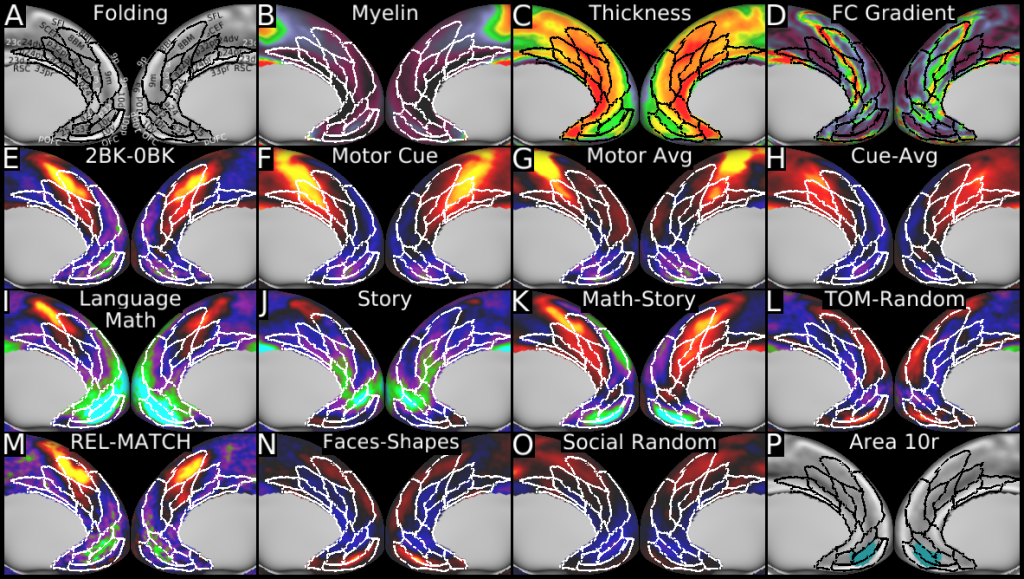
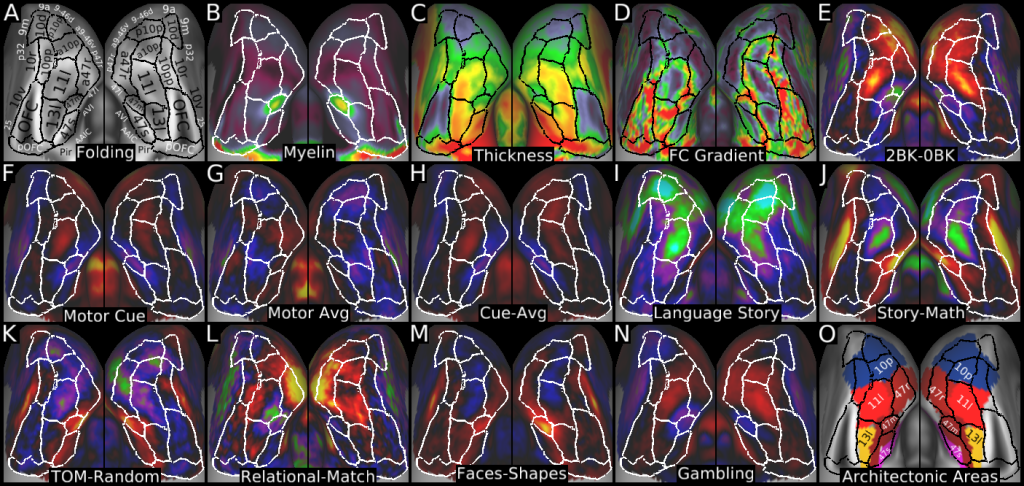
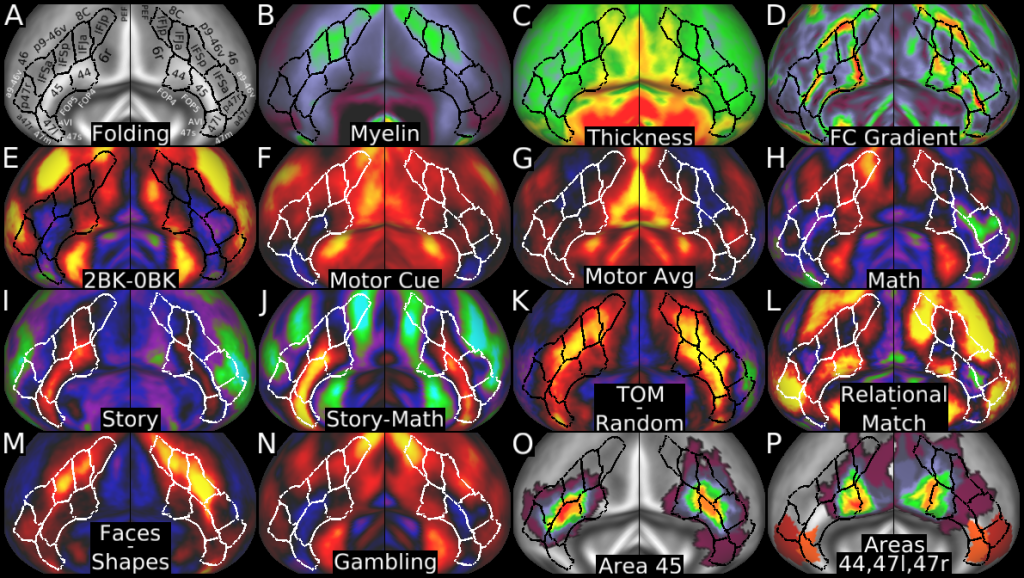
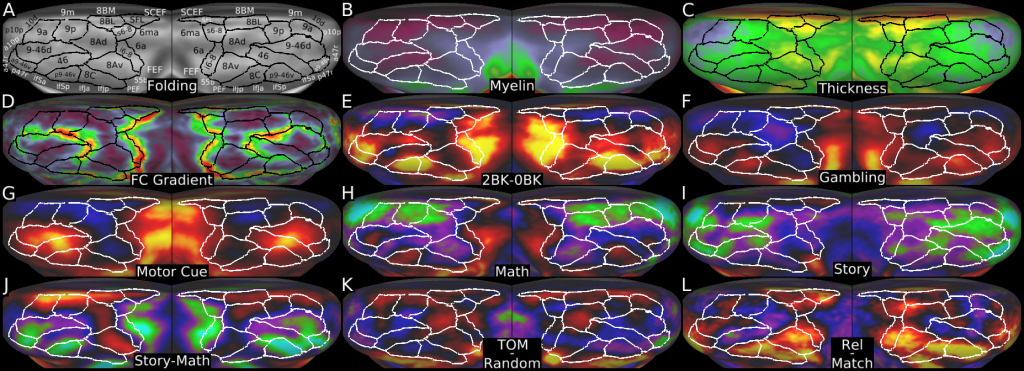
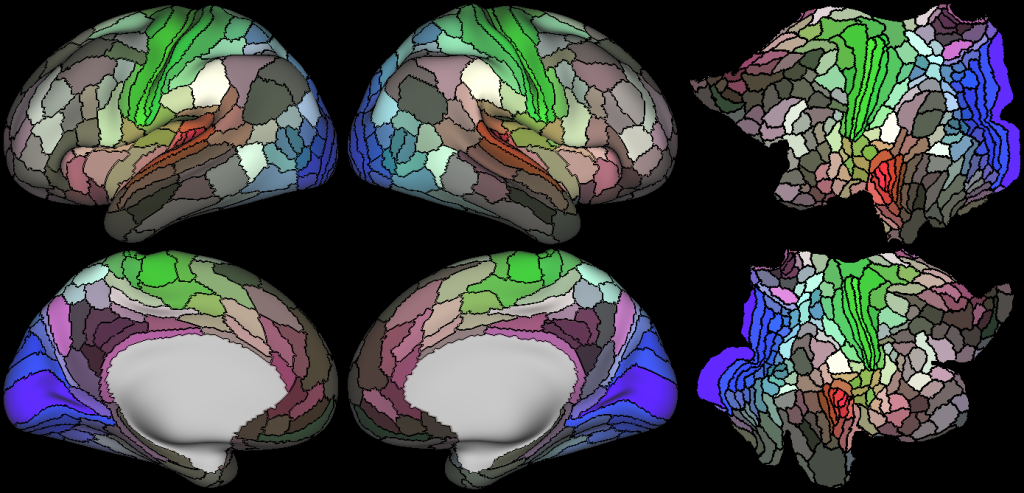
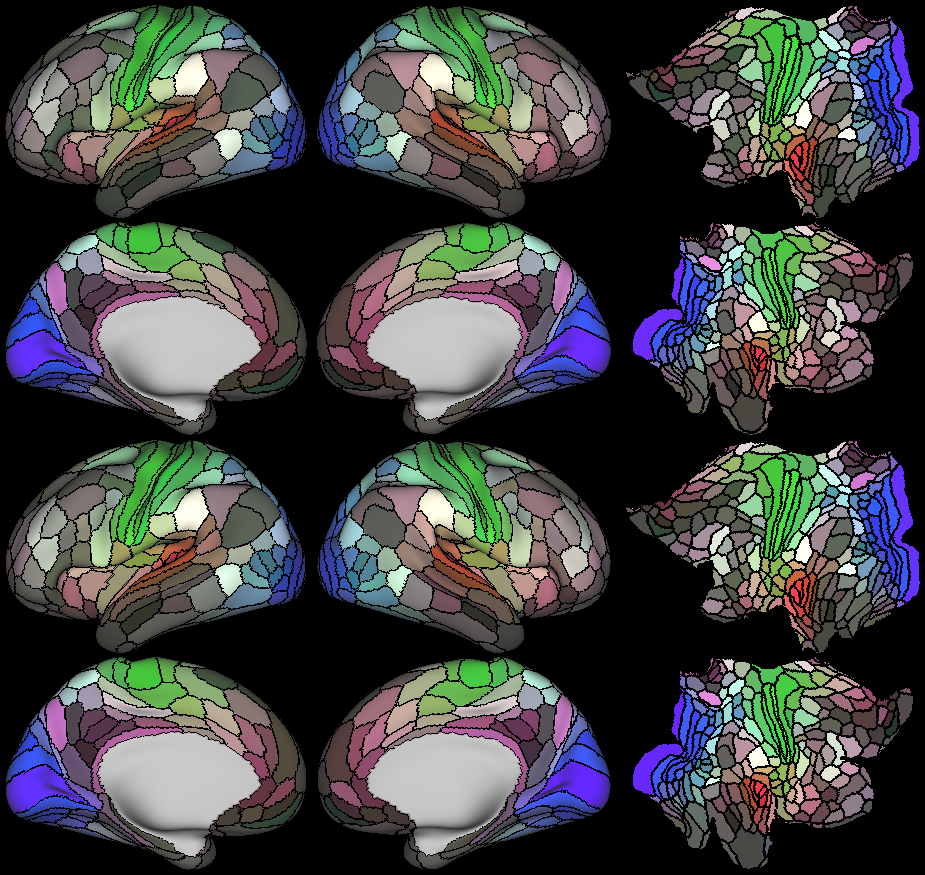
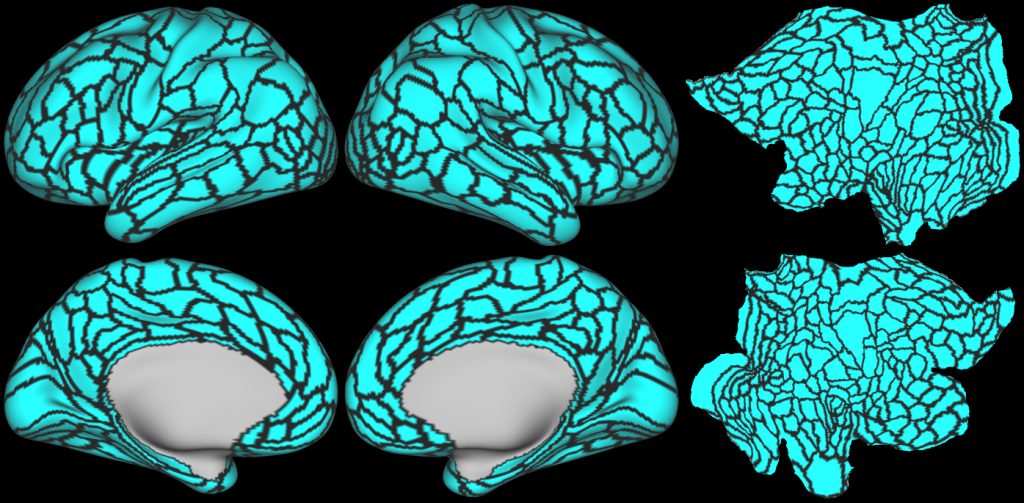
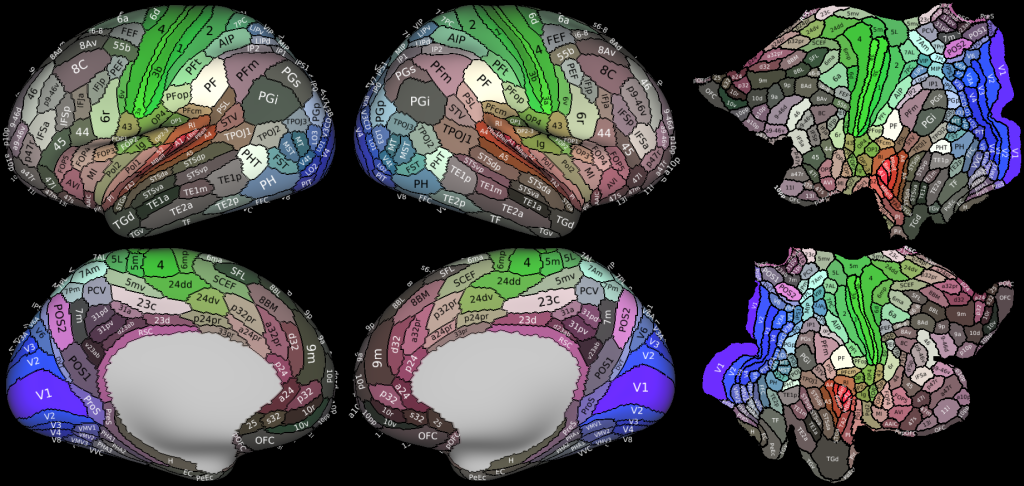
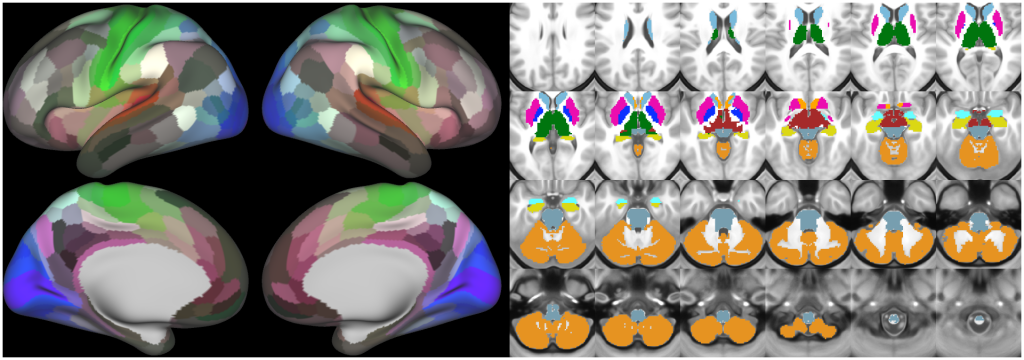
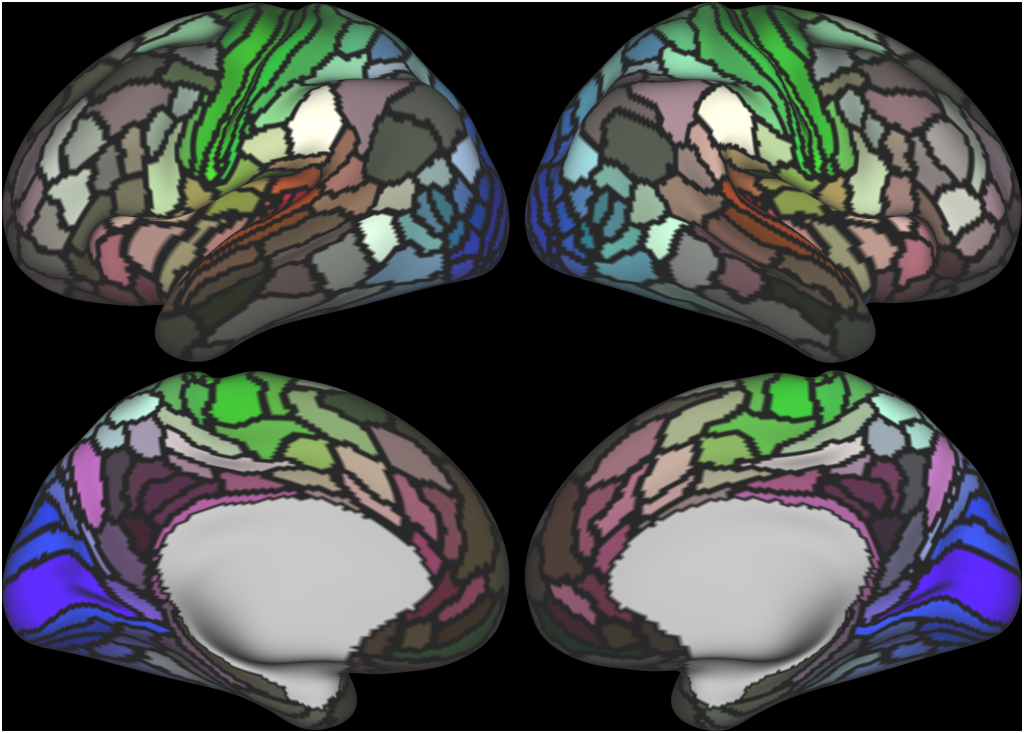
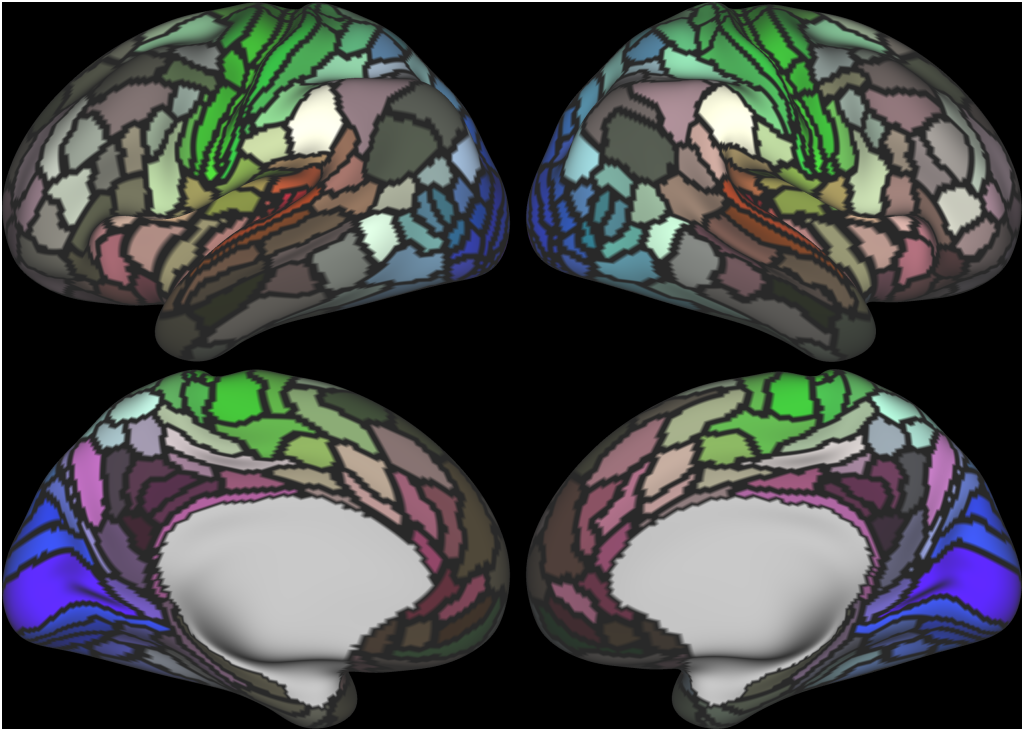
Understanding the amazingly complex human cerebral cortex requires a map (or parcellation) of its major subdivisions, known as cortical areas. Making an accurate areal map has been a century-old objective in neuroscience. Using multimodal magnetic resonance images from the Human Connectome Project (HCP) and an objective semi-automated neuroanatomical approach, we delineated 180 areas per hemisphere bounded by sharp changes in cortical architecture, function, connectivity, and/or topography in a precisely aligned group average of 210 healthy young adults. We characterized 97 new areas and 83 areas previously reported using post-mortem microscopy or other specialized studyspecific approaches. To enable automated delineation and identification of these areas in new HCP subjects and in future studies, we trained a machine-learning classifier to recognize the multi-modal ‘fingerprint’ of each cortical area. This classifier detected the presence of 96.6% of the cortical areas in new subjects, replicated the group parcellation, and could correctly locate areas in individuals with atypical parcellations. The freely available parcellation and classifier will enable substantially improved neuroanatomical precision for studies of the structural and functional organization of human cerebral cortex and its variation across individuals and in development, aging, and disease.
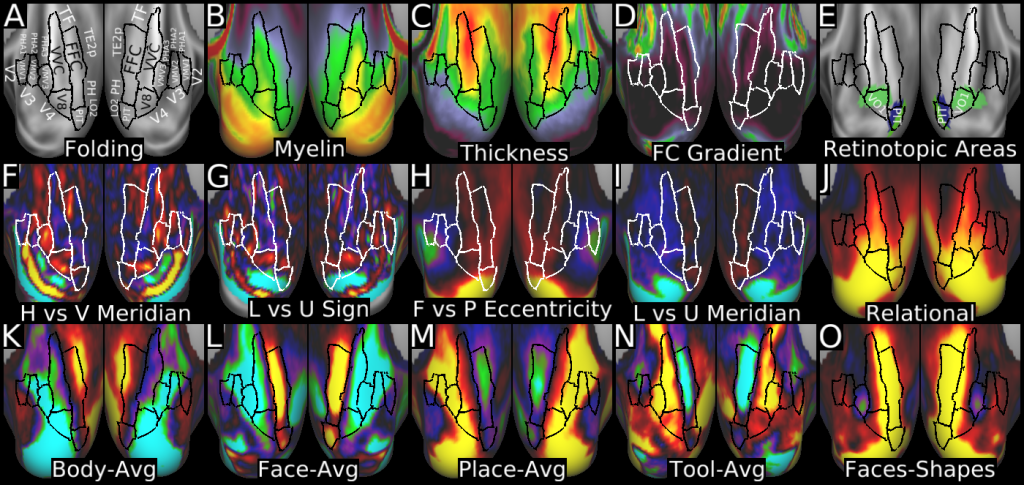
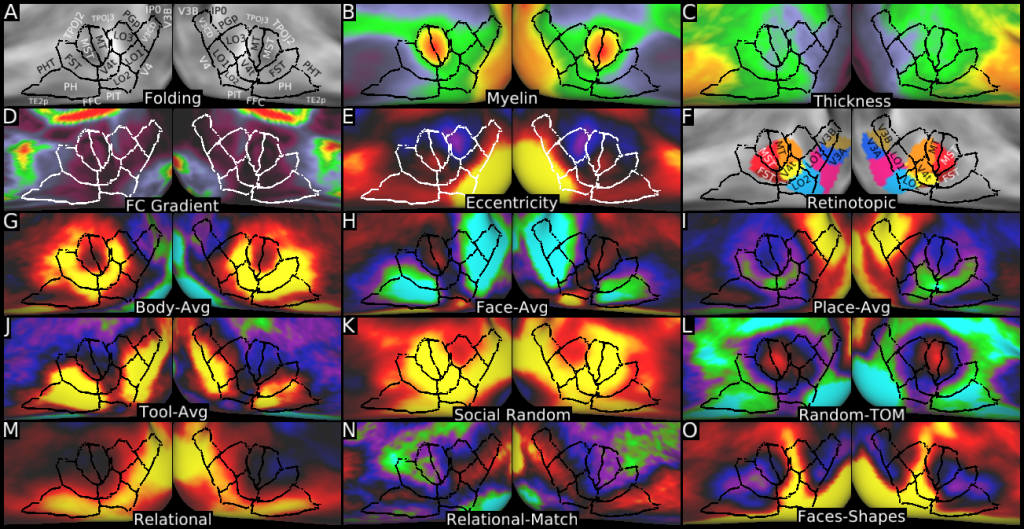
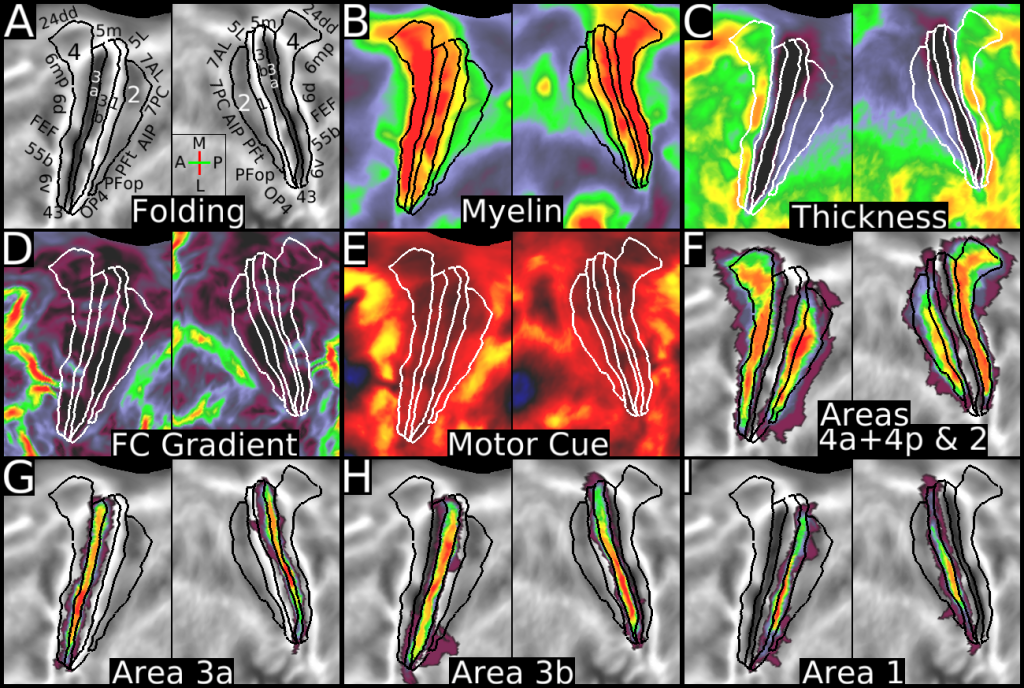
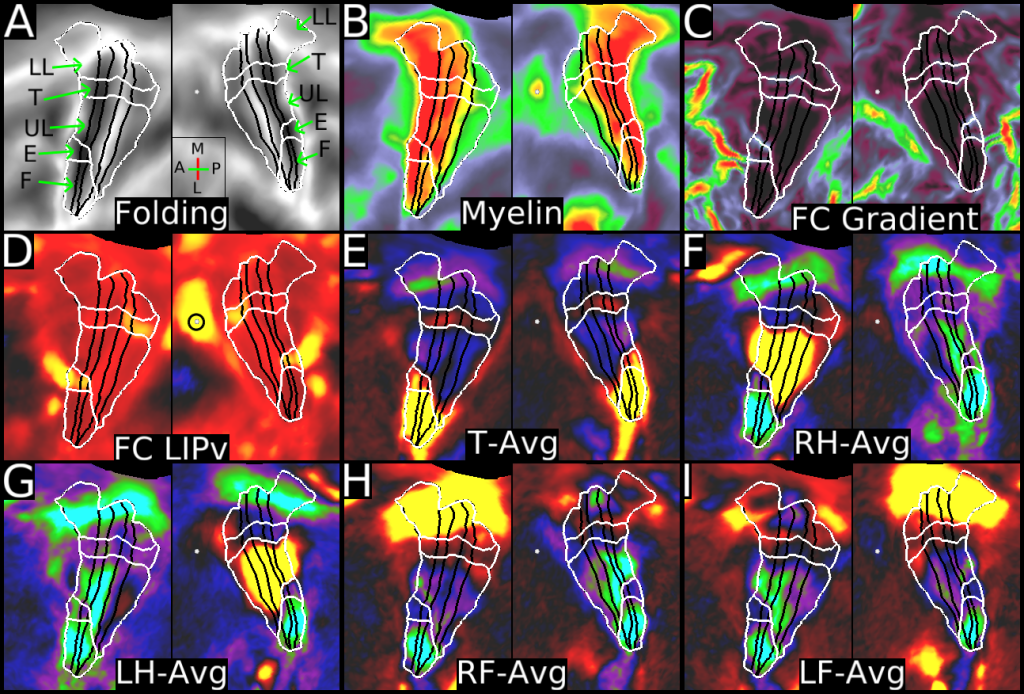
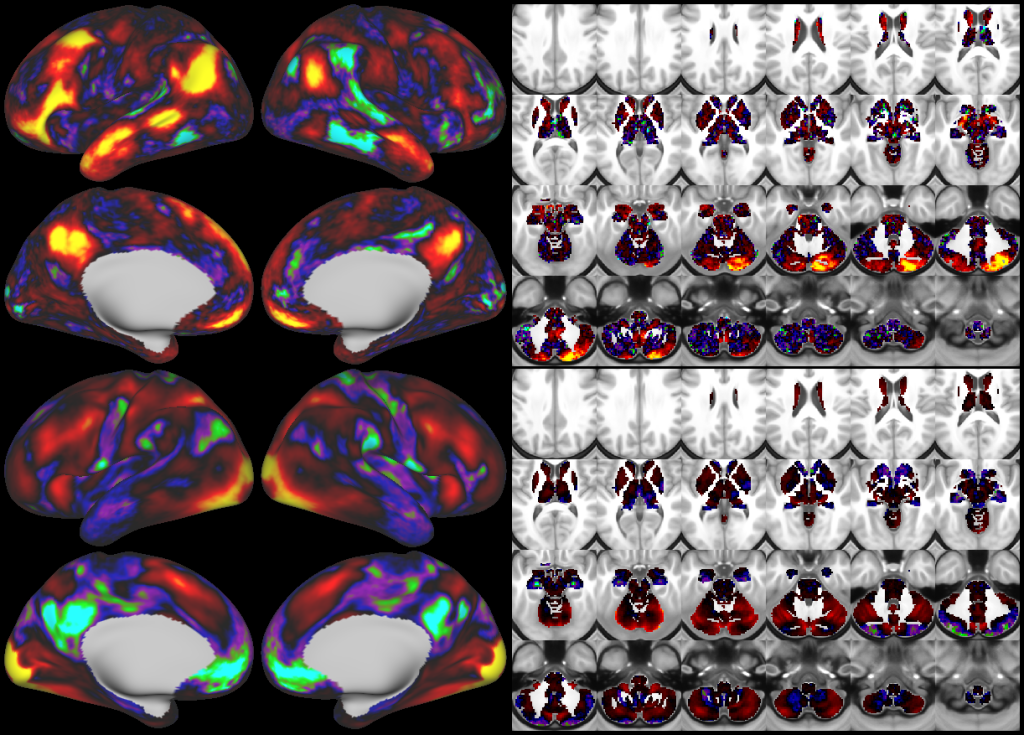
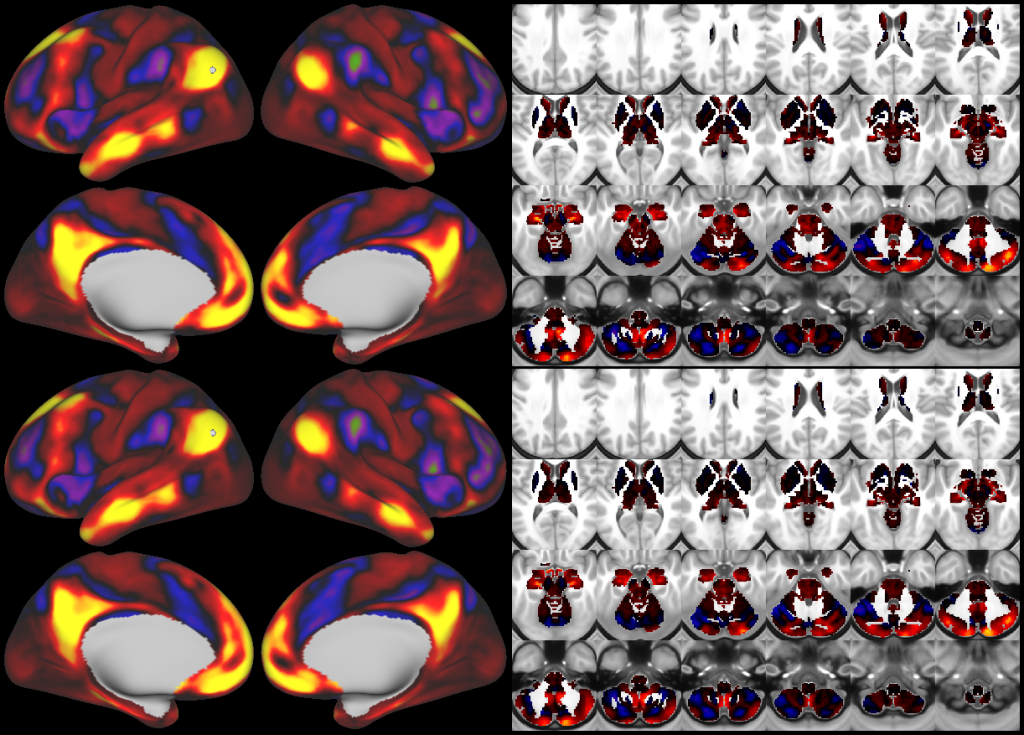
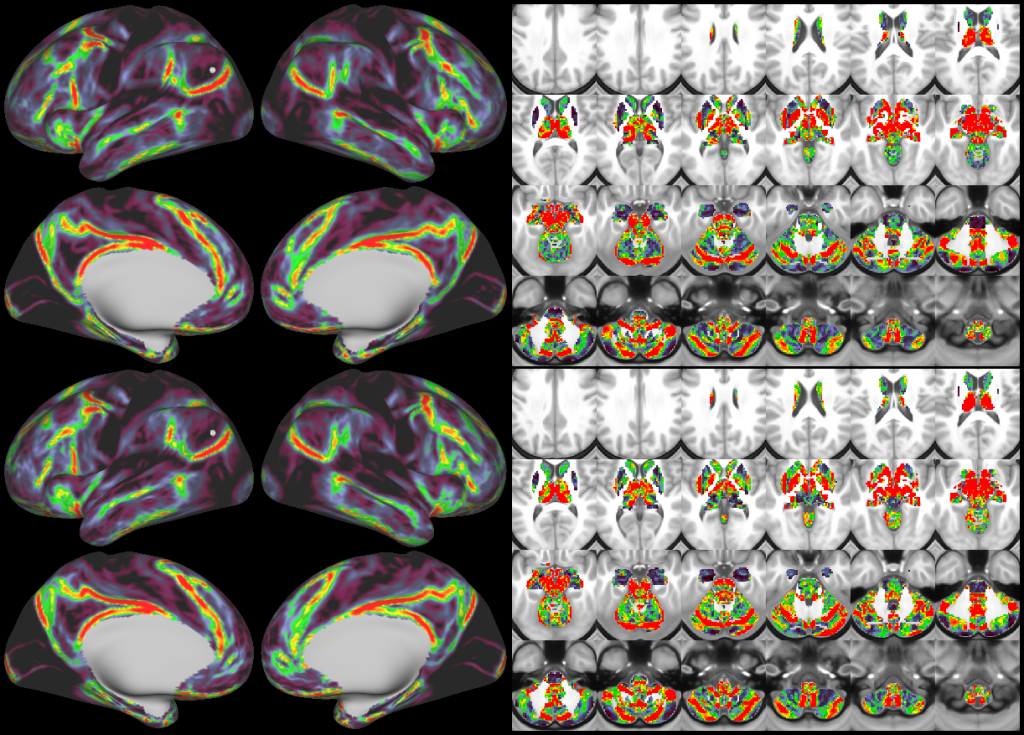
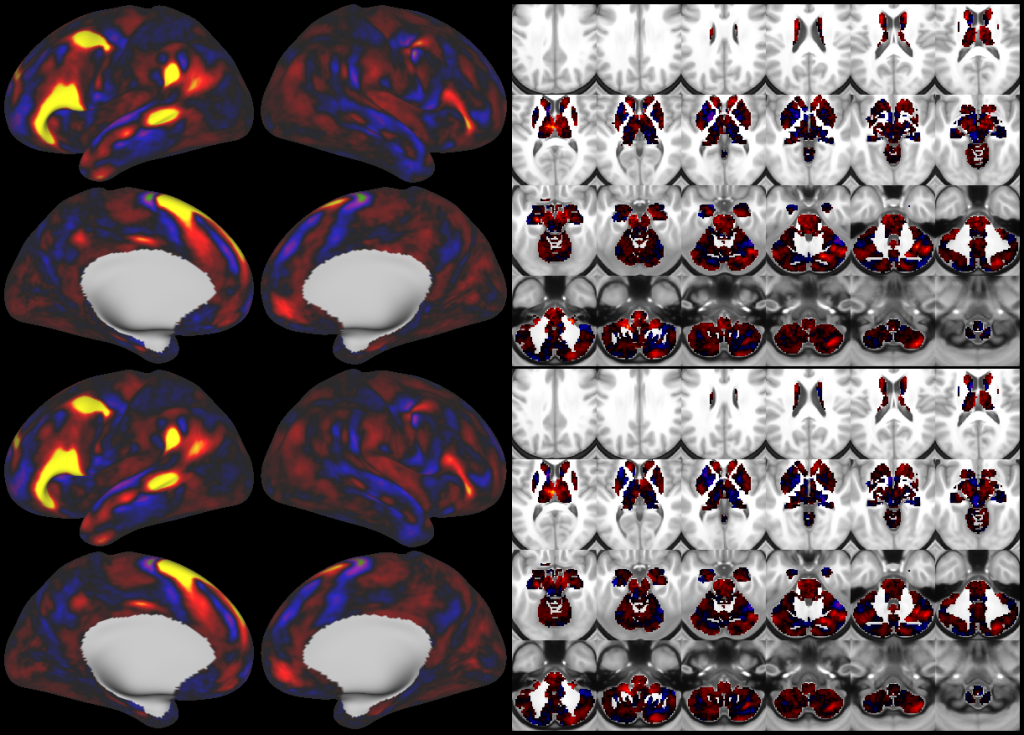
This publication is organized into 4 documents: (1) A main text, (2) A supplementary results and discussion, (3) A supplementary methods, and (4) A supplementary neuroanatomical results. Each of these documents has its own scene file associated with it. In addition we provide a scene file (5) that contains the main study datasets. This study dataset scene file is intended for those wanting to do follow up analyses using the data shared by this publication, whereas the other scene files are intended for those who want to explore the data shown in each figure interactively. Several people requested scene file (6) which shows all of the areas labled. Note that Connectome Workbench Version 1.5.0 or greater is required to view these scenes. See the help at the top right of the screen for a link to the latest Connectome Workbench.
For questions about this study, please post on the HCP-Users Mailing List:
https://www.humanconnectome.org/contact/hcp-users-request.php
The files included in this study are CIFTI, GIFTI, and NIFTI and the parcellation is in CIFTI format. CIFTI files are not viewable in a standard NIFTI volume file viewer, but are viewable in Connectome Workbench and can be analyzed in Matlab, FSL's PALM, or the next version of FSL. Many have chosen to use the 210V Maximum Probability Map (MPM) parcellation (HCP_PhaseTwo / Q1-Q6_RelatedValidation210 / MNINonLinear / fsaverage_LR32k / Q1-Q6_RelatedValidation210.CorticalAreas_dil_Final_Final_Areas_Group_Colors.32k_fs_LR.dlabel.nii) as likely being the most similar to a future 1071 MPM that will be produced by the HCP. This file is available here: https://balsa.wustl.edu/file/show/3VLx or here with subcortical structures: https://balsa.wustl.edu/file/87B9N. We have also made available colorized versions of the original (non-MPM) parcellations for cortical areas, cortical areas plus sensorimotor subregions, and cortical areas plus sensorimotor subareas: https://balsa.wustl.edu/sceneFile/mlwnX.
ABSTRACT:
Understanding the amazingly complex human cerebral cortex requires a map (or parcellation) of its major subdivisions, known as cortical areas. Making an accurate areal map has been a century-old objective in neuroscience. Using multimodal magnetic resonance images from the Human Connectome Project (HCP) and an objective semi-automated neuroanatomical approach, we delineated 180 areas per hemisphere bounded by sharp changes in cortical architecture, function, connectivity, and/or topography in a precisely aligned group average of 210 healthy young adults. We characterized 97 new areas and 83 areas previously reported using post-mortem microscopy or other specialized studyspecific approaches. To enable automated delineation and identification of these areas in new HCP subjects and in future studies, we trained a machine-learning classifier to recognize the multi-modal ‘fingerprint’ of each cortical area. This classifier detected the presence of 96.6% of the cortical areas in new subjects, replicated the group parcellation, and could correctly locate areas in individuals with atypical parcellations. The freely available parcellation and classifier will enable substantially improved neuroanatomical precision for studies of the structural and functional organization of human cerebral cortex and its variation across individuals and in development, aging, and disease.
PUBLICATION:
Nature
- DOI:
10.1038/nature18933
- PMID:
27437579
- Matthew F. Glasser
- Timothy S. Coalson
- Emma C. Robinson
- Carl D. Hacker
- John Harwell
- Essa Yacoub
- Kamil Ugurbil
- Jesper Andersson
- Christian F Beckmann
- Mark Jenkinson
- Stephen M. Smith
- David C. Van Essen
-
Glasser_et_al_2016_HCP_MMP1.0_1_MainText.scene
SCENES: -
Glasser_et_al_2016_HCP_MMP1.0_2_SupplementaryResultsAndDiscussion.scene
SCENES: -
Glasser_et_al_2016_HCP_MMP1.0_3_SupplementaryMethods.scene
SCENES: -
Glasser_et_al_2016_HCP_MMP1.0_4_SupplementaryNeuroanatomicalResults.scene
SCENES: -
Glasser_et_al_2016_HCP_MMP1.0_5_StudyDataset.scene
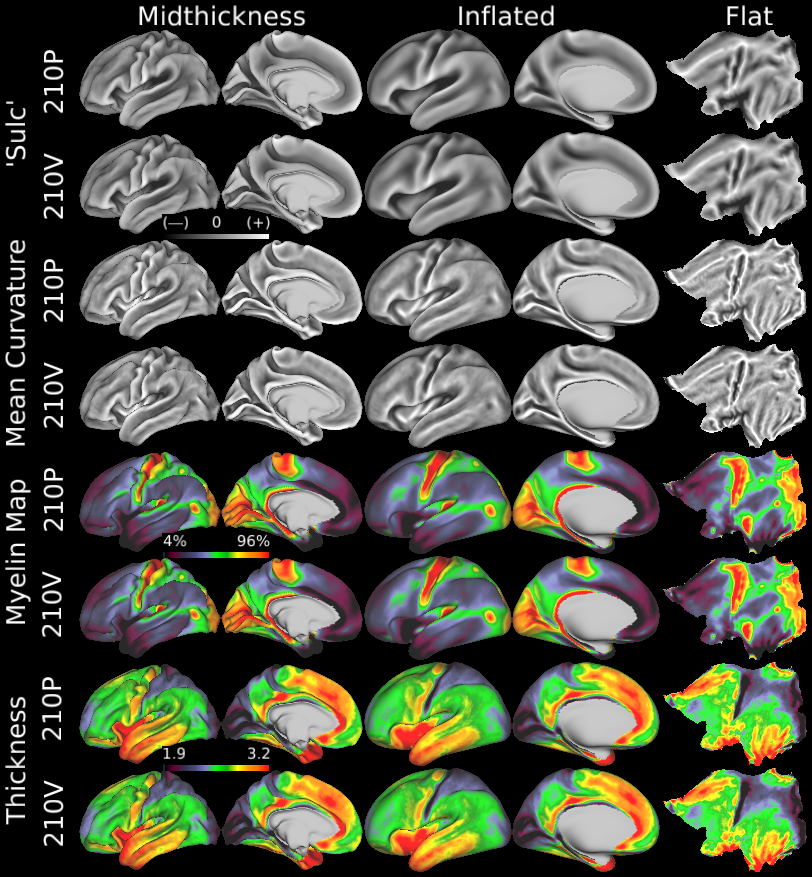
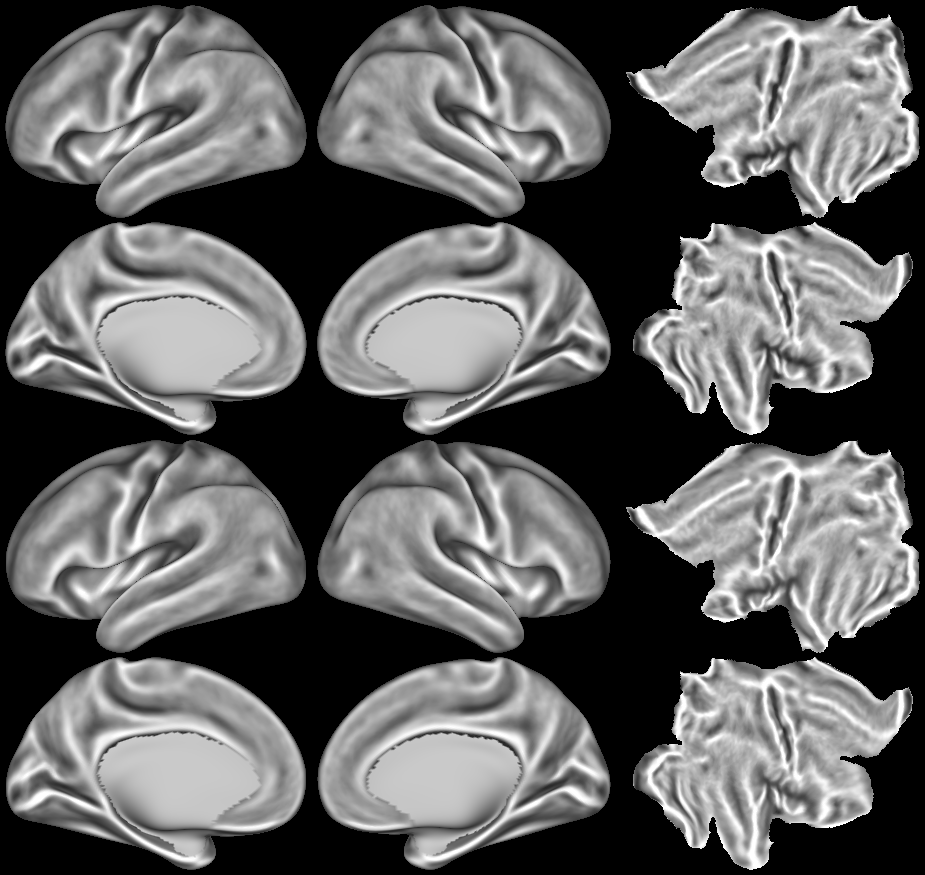
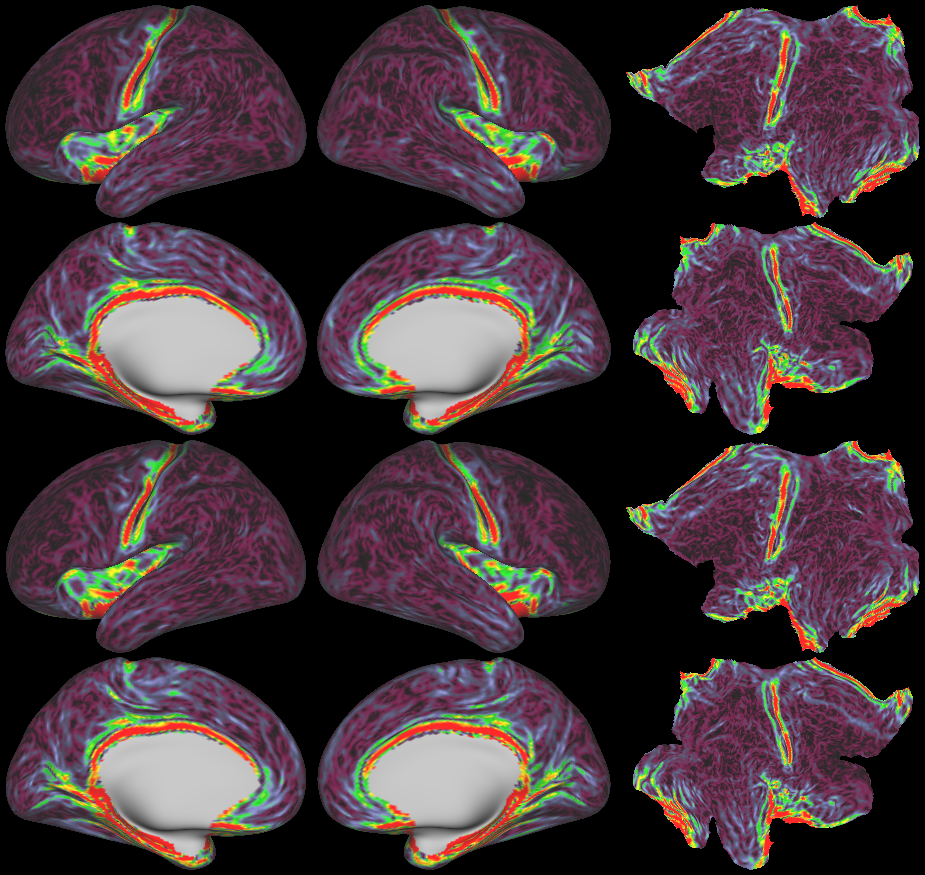
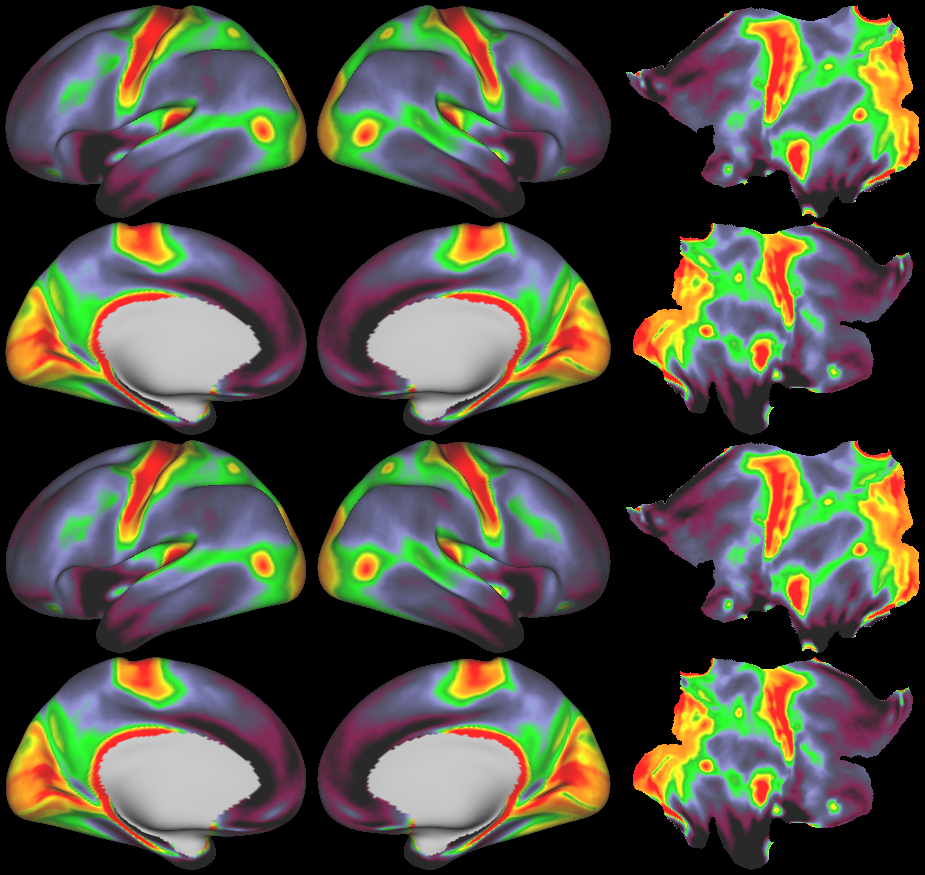
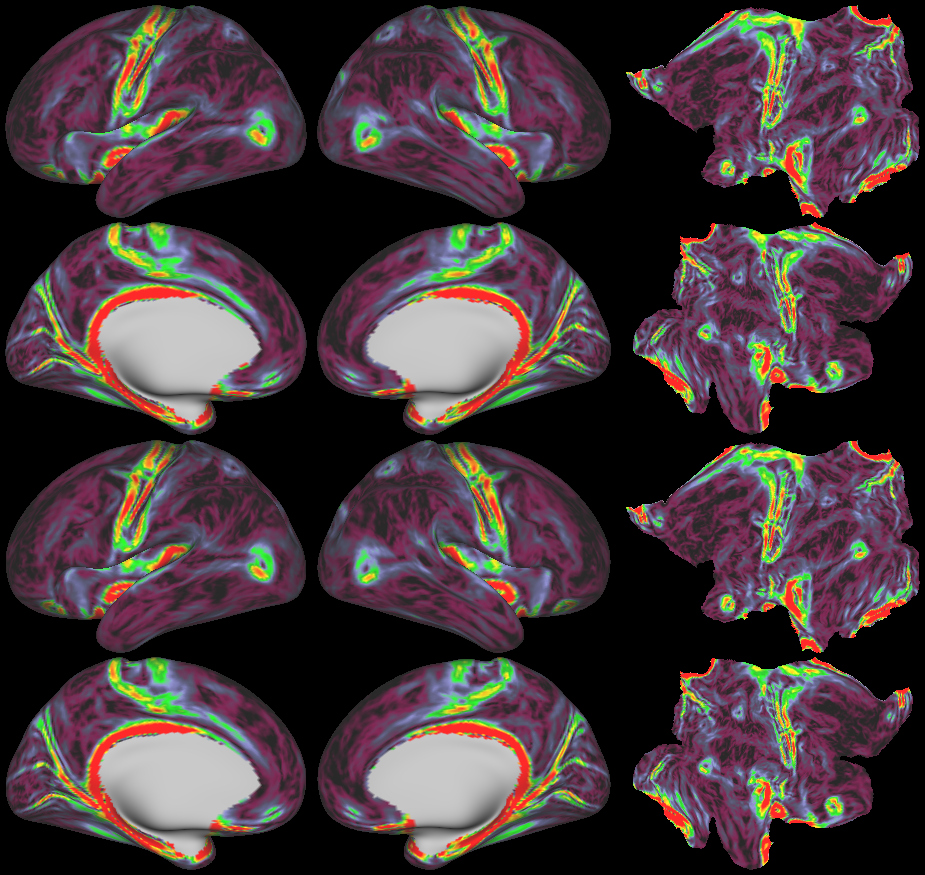
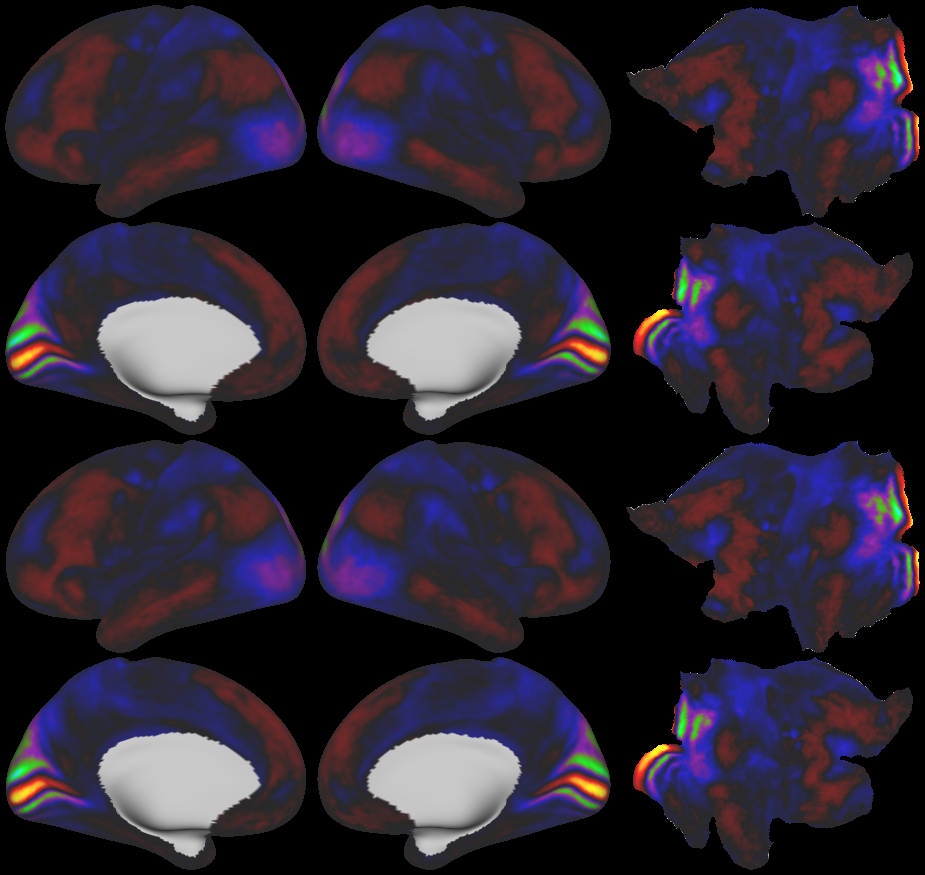
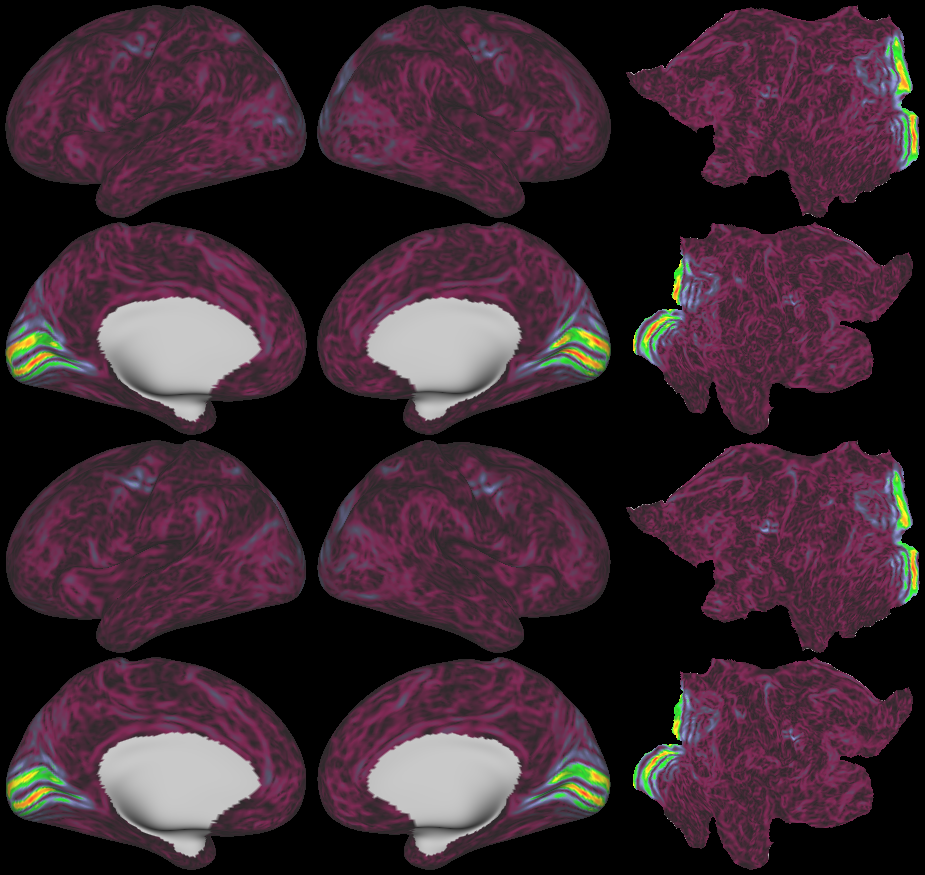
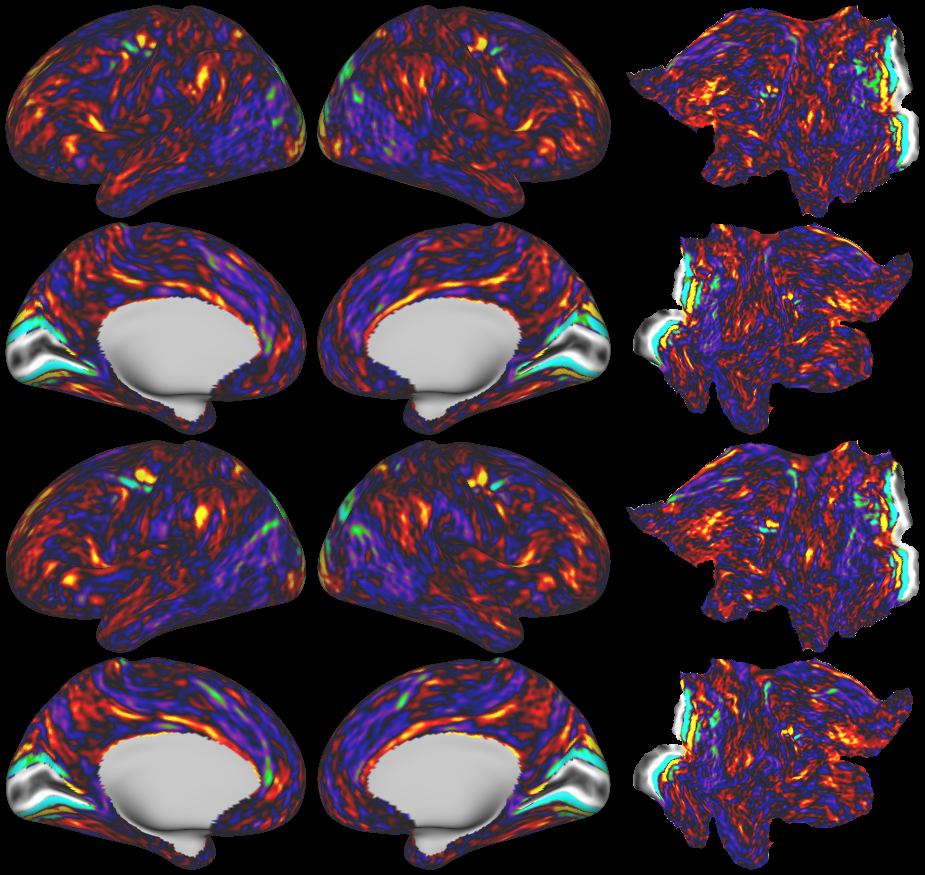
SCENES:- Cortical Folding
- Cortical Thickness
- Cortical Thickness Gradient
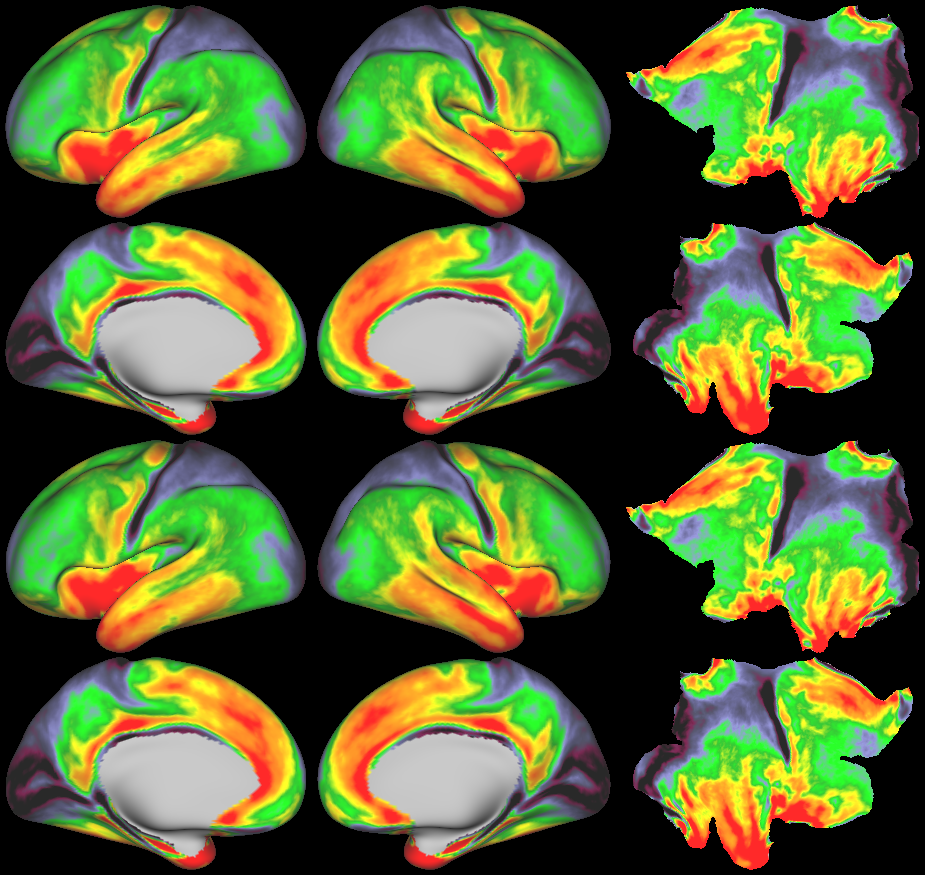
- Cortical Myelin Maps
- Cortical Myelin Gradient
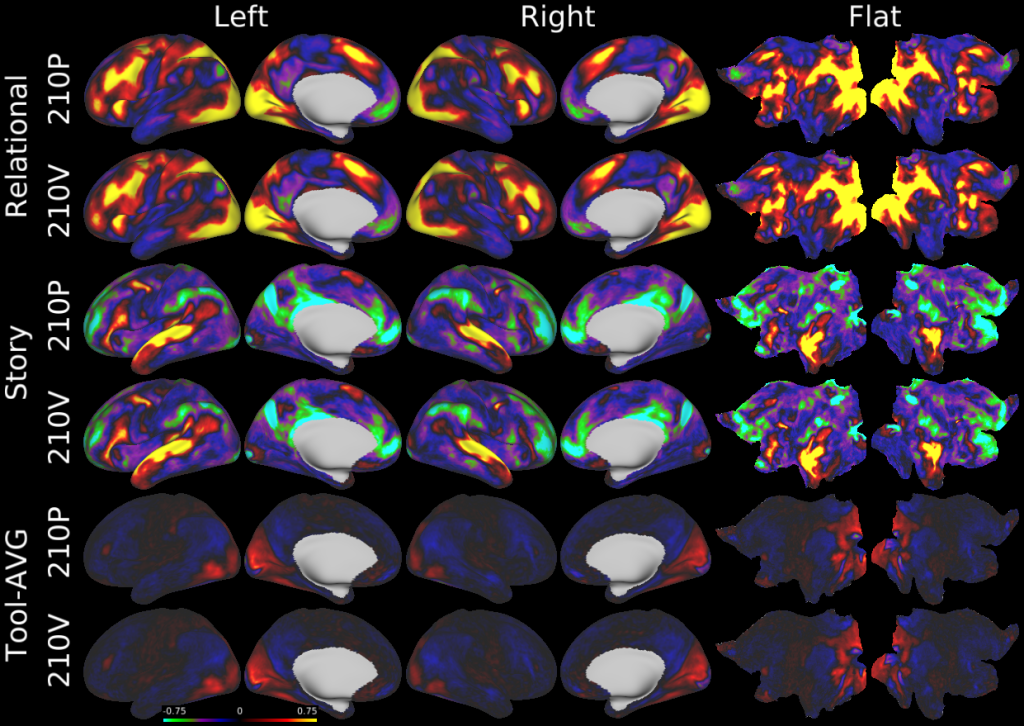
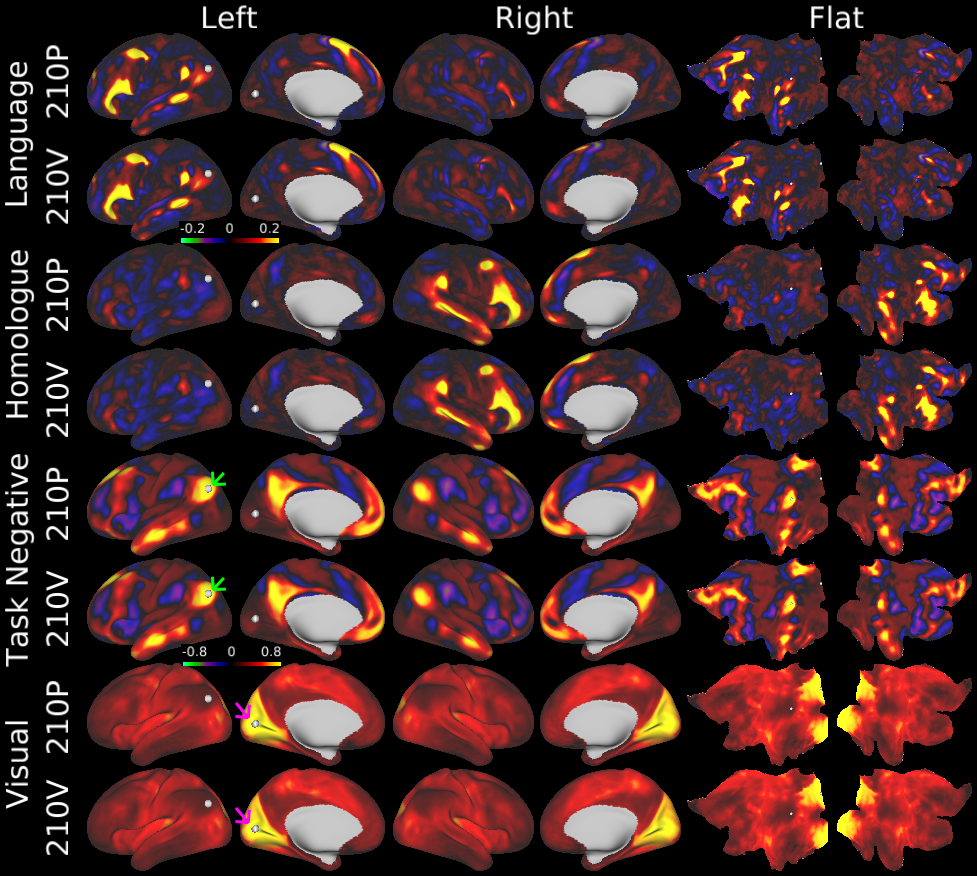
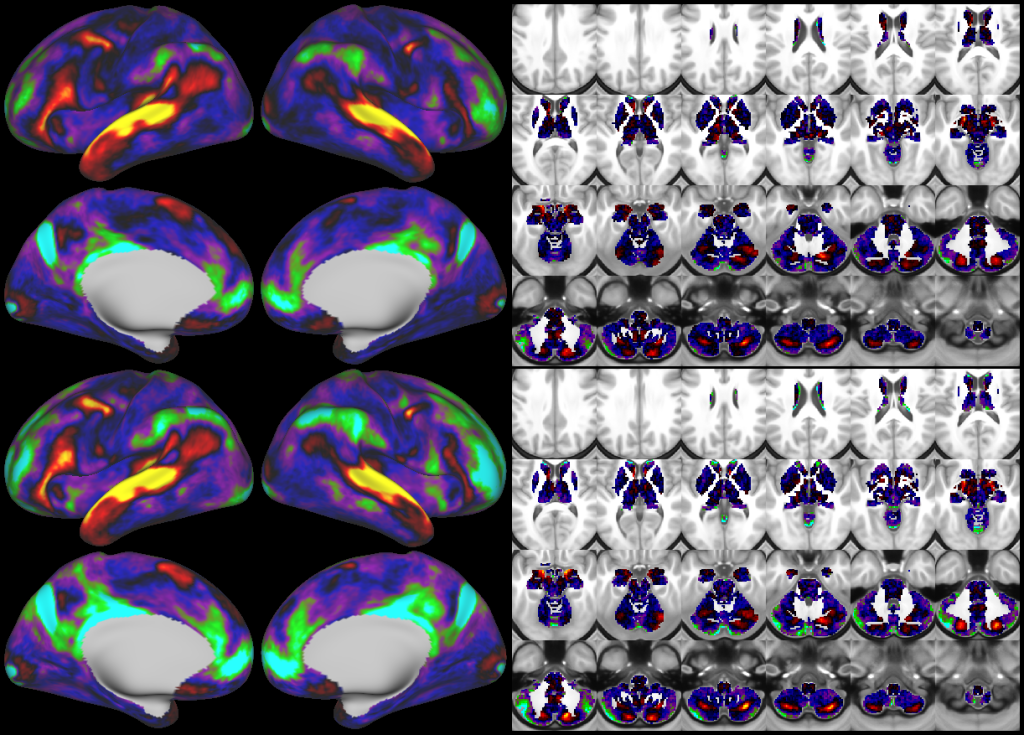
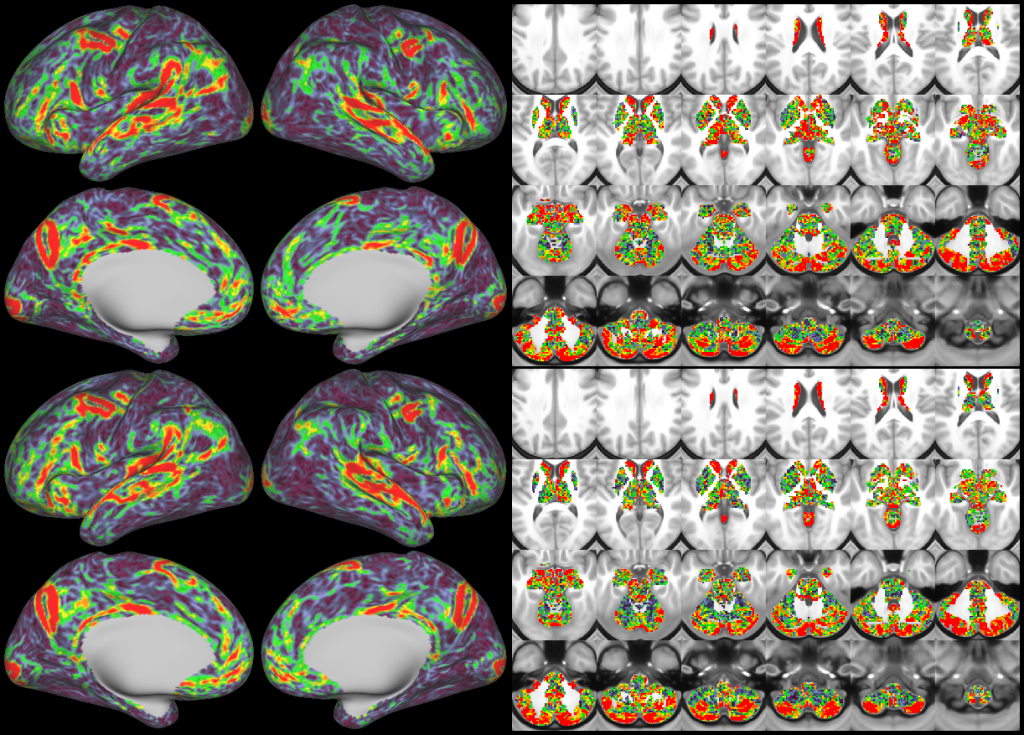
- Task fMRI Beta Maps
- Task fMRI Gradient Maps
- d=20+1 tfMRI Contrast ICA Dimensionality Reduction
- Dynamic Dense Functional Connectomes
- Mean Functional Connectivity Gradients
- d=137 Resting State Networks (RSNs)
- Visuotopic Maps
- Visuotopic Gradients
- Visuotopic Sign Maps
- Original Semiautomated Parcellation
- Probabilistic Areas
- Group MPM Parcellations
- Original Semiautomated Parcellation with Somatosensory Motor Sub Regions
-
Glasser_et_al_2016_HCP_MMP1.0_6_AllAreasMap.scene
SCENES: -
Glasser_et_al_2016_HCP_MMP1.0_7_CorticalPlusSubcortical.scene
SCENES: -
Glasser_et_al_2016_HCP_MMP1.0_8_CorticalAreasPlusSubRegionsPlusSubAreas.scene
SCENES: