FULL TITLE:
Modelling white matter in gyral blades as a continuous vector field
SPECIES:
Human
DESCRIPTION:
Contains scene files to recreate the figures in the paper.
Explanation for the directory structure. At the top level there are 3 directories:
- scenes: contains the scene files
- input: contains any data downloaded from the HCP Q1200 datasets
- data: contains the data generated in this study
Within each of these directories, the files might be hidden in lots of sub-directories, however they should be fairly easy to find as long as you remember that any results generated using tracking from/to the white/grey-matter boundary will be in a directory labeled "white", while those tracking from/to the new deep/gyral white matter interface will be labeled "deformed".
ABSTRACT:
Many brain imaging studies aim to measure structural connectivity with diffusion tractography. However, biases in tractography data, particularly near the boundary between white matter and cortical grey matter can limit the accuracy of such studies. When seeding from the white matter, streamlines tend to travel parallel to the convoluted cortical surface, largely avoiding sulcal fundi and terminating preferentially on gyral crowns. When seeding from the cortical grey matter, streamlines generally run near the cortical surface until reaching deep white matter. These so-called “gyral biases” limit the accuracy and effective resolution of cortical structural connectivity profiles estimated by tractography algorithms, and they do not reflect the expected distributions of axonal densities seen in invasive tracer studies or stains of myelinated fibres. We propose an algorithm that concurrently models fibre density and orientation using a divergence-free vector field within gyral blades to encourage an anatomically-justified streamline density distribution along the cortical white/grey-matter boundary while maintaining alignment with the diffusion MRI estimated fibre orientations. Using in vivo data from the Human Connectome Project, we show that this algorithm reduces tractography biases. We compare the structural connectomes to functional connectomes from resting-state fMRI, showing that our model improves cross-modal agreement. Finally, we find that after parcellation the changes in the structural connectome are very minor with slightly improved interhemispheric connections (i.e, more homotopic connectivity) and slightly worse intrahemispheric connections when compared to tracers.
- Michiel Cottaar
- Matteo Bastiani
- Nikhil Boddu
- Matthew Glasser
- Suzanne Haber
- David C. Van Essen
- Stamatios N. Sotiropoulos
- Saad Jbabdi
- Washington University in St. Louis
- University of Nottingham
- University of Oxford
-
cost_func.scene
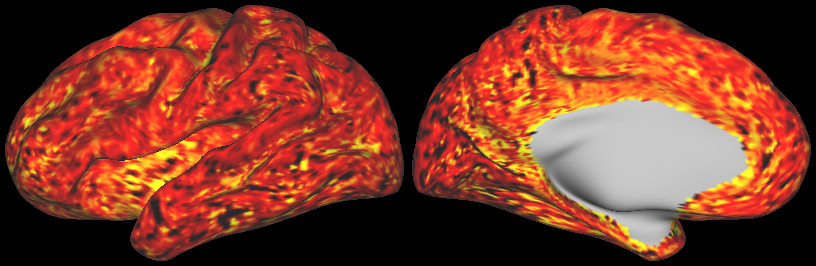
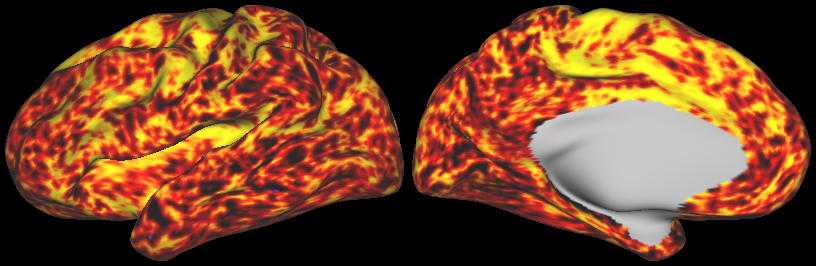
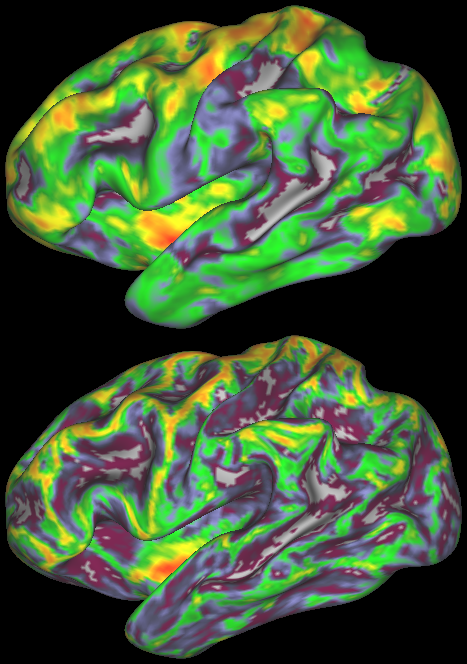
DESCRIPTION: Figure 5: Vector field evaluated at the cortical surface (white/grey-matter boundary, midcortical surface, and pial surface).
SCENES: -
track_gyral_wm.scene
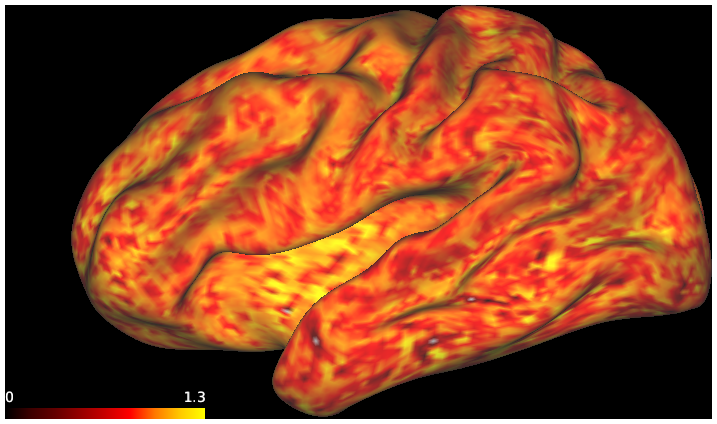
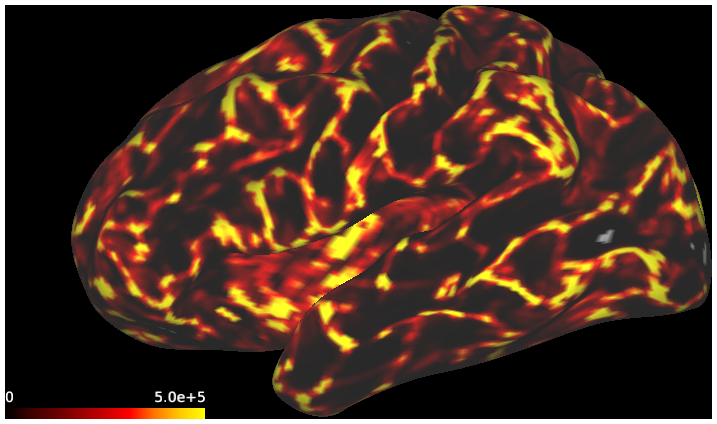
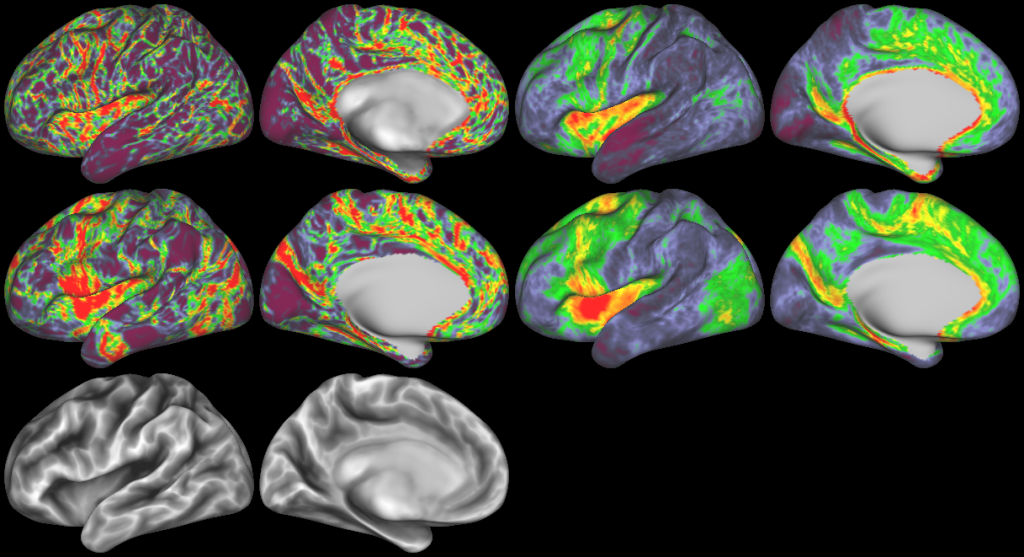
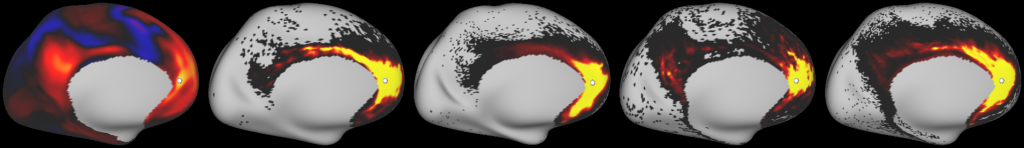
DESCRIPTION: Figure 7: surface streamline density from either the vector field or classical tractography through the gyral blade white matter
SCENES: -
dconn_log_density.scene
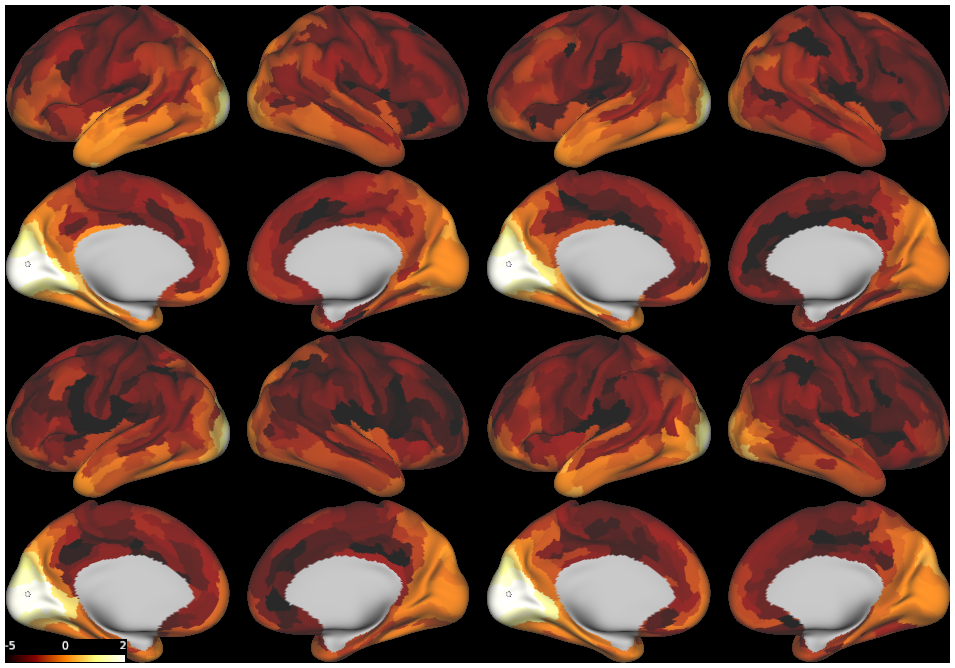
DESCRIPTION: Figure 8: density (log-scale) of streamline termination points with (top) and without (bottom) adopting the vector field model
SCENES: -
gradients2_upload.scene
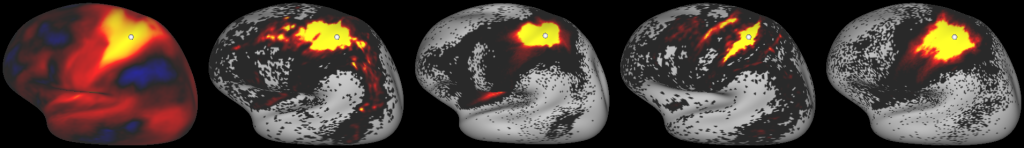
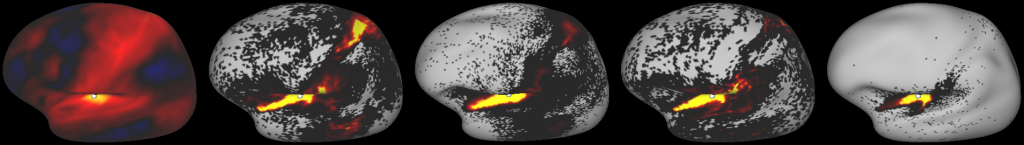
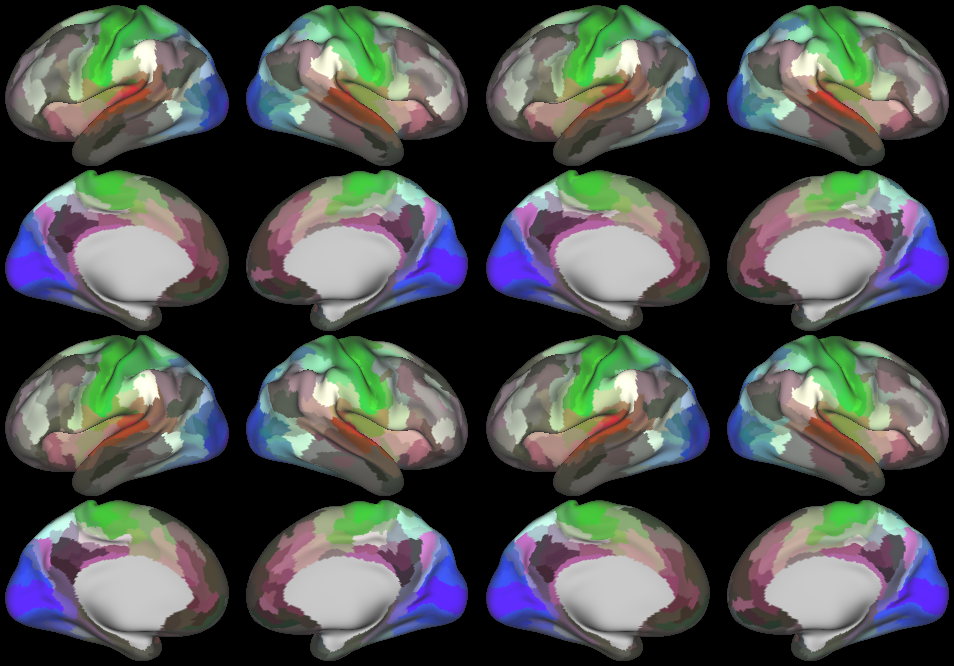
DESCRIPTION: Figure 9: Gradients in connectivity profile when seeding at the cortical surface
SCENES: -
fconn_comparison.scene
DESCRIPTION: Figure 10: Comparison of the functional and structural connectivity
SCENES: -
mm_parc.scene
DESCRIPTION: Figure S2: parcellated structural connectome
SCENES: