FULL TITLE:
Dynamic patterns of cortical expansion during folding of the preterm human brain
SPECIES:
Human
DESCRIPTION:
The human brain exhibits complex folding patterns that emerge during the third trimester of fetal development. Despite decades of study, the processes that produce normal and abnormal folding remain unresolved, although the relatively rapid tangential expansion of the cortex has emerged as a driving factor. Accurate and precise measurement of cortical growth patterns during the period of folding has remained elusive. Here, we illuminate the spatiotemporal dynamics of cortical expansion by analyzing MRI-derived surfaces of preterm infant brains, using a novel strain energy minimization approach.
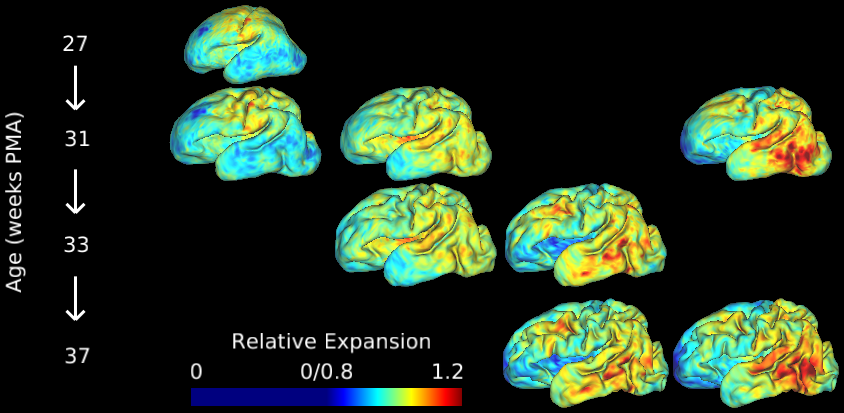
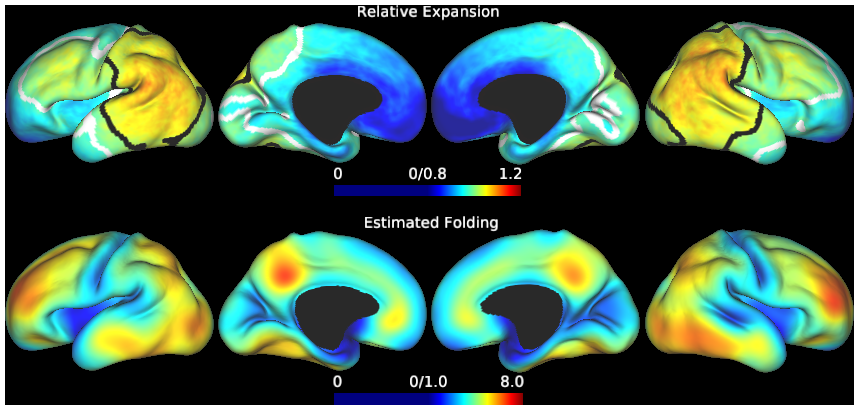
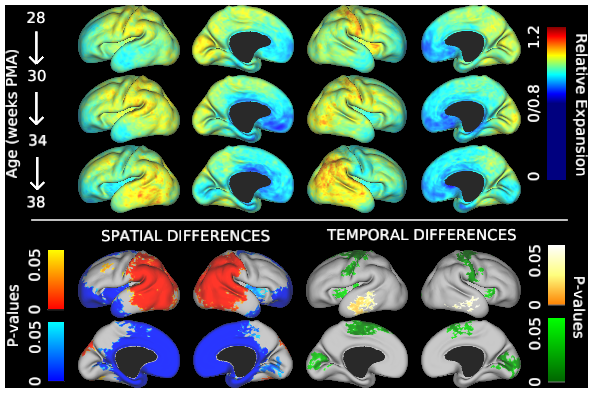
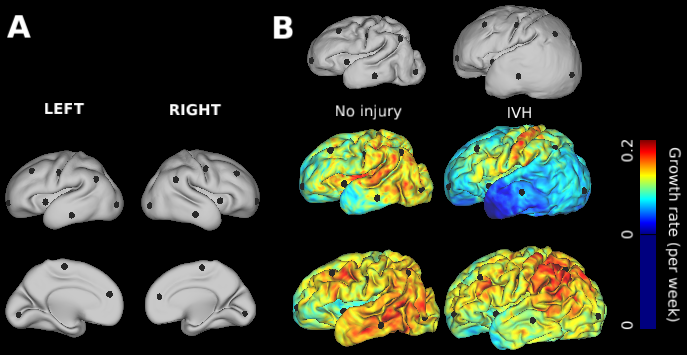
This study contains cortical growth maps over the course of preterm development, measured from human mid-thickness surfaces. Cortical expansion was calculated from local deformations after registration with anatomically-constrained Multimodal Surface Matching (aMSM, Robinson et al. Neuroimage 2017). Each scene corresponds to a figure from the main Results in Garcia et al. PNAS 2018. In addition to summary data displayed in Figs. 4-6 (group averages, statistically significant regions), individual maps are also made available in the corresponding scene.
The zipped folder "MATLABsimulation" contains a simple example of strain energy minimization between two surfaces. It requires two closed surfaces (on which to minimize strain energy) as well as a spherical surface to which they can be mapped, all with the same number of vertices and connections. This code does not match landmarks, so surfaces should first be brought into approximate register for best results. The main code (relax.m) follows the approach outlined in Knutsen et al. (2010), replacing the steps requiring transition from MATLAB to Comsol Multiphysics (solving equations of motion). Note: This code calls MATLAB codes developed in Knutsen et al. (2010) (included) and codes for importing GIfTI surface files available at https://www.artefact.tk/software/matlab/gifti/
ABSTRACT:
During the third trimester of human brain development, the cerebral cortex undergoes dramatic surface expansion and folding. Physical models suggest that relatively rapid growth of the cortical gray matter helps drive this folding, and structural data suggests that growth may vary in both space (by region on the cortical surface) and time. In this study, we propose a new method to estimate local growth from sequential cortical reconstructions. Using anatomically-constrained Multimodal Surface Matching (aMSM), we obtain accurate, physically-guided point correspondence between younger and older cortical reconstructions of the same individual. From each pair of surfaces, we calculate continuous, smooth maps of cortical expansion with unprecedented precision. By considering 30 preterm infants scanned 2-4 times during the period of rapid cortical expansion (28 to 38 weeks postmenstrual age), we observe significant regional differences in growth across the cortical surface that are consistent with the emergence of new folds. Furthermore, these growth patterns shift over the course of development, with non-injured subjects following a highly consistent trajectory. This information provides a detailed picture of dynamic changes in cortical growth, connecting what is known about patterns of development at the microscopic (cellular) and macroscopic (folding) scales. Since our method provides specific growth maps for individual brains, we are also able to detect alterations due to injury. This fully-automated surface analysis, based on tools freely available to the brain mapping community, may also serve as a useful approach for future studies of abnormal growth due to genetic disorders, injury, or other environmental variables.
PUBLICATION:
Proceedings of the National Academy of Sciences
- DOI:
10.1073/pnas.1715451115
- Kara E. Garcia
- Emma C. Robinson
- Dimitrios Alexopoulos
- Donna L. Dierker
- Matthew F. Glasser
- Timothy S. Coalson
- Cynthia M. Ortinau
- Daniel Rueckert
- Larry A. Taber
- David C. Van Essen
- Cynthia E. Rogers
- Christopher D. Smyser
- Philip V. Bayly
- Imperial College London
- Washington University in St. Louis
- Kings College London