FULL TITLE:
Cerebral Cortical Folding, Parcellation, and Connectivity in Humans, Nonhuman Primates, and Mice
SPECIES:
Human, Macaque, Chimpanzee
ABSTRACT:
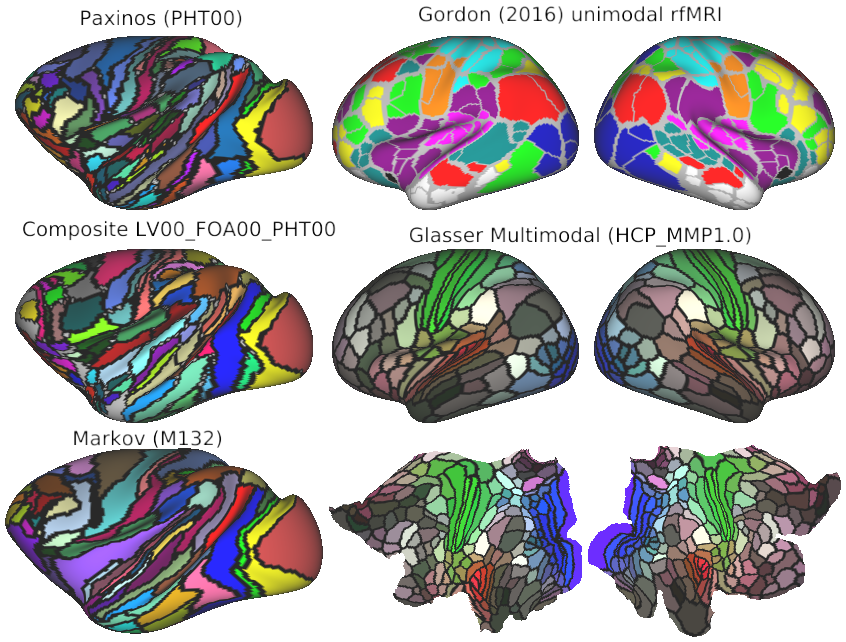
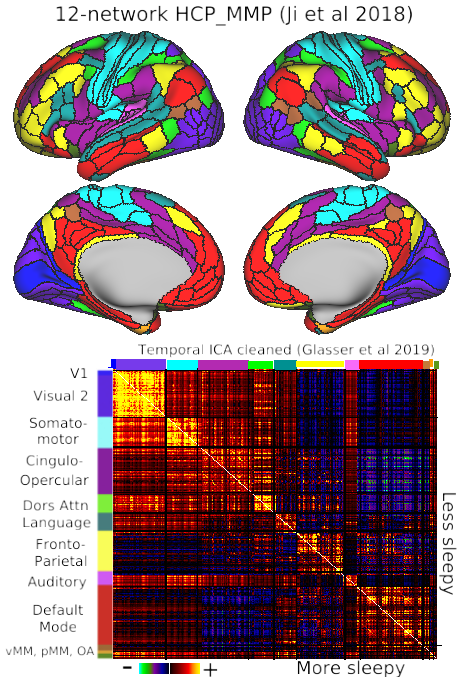
Advances in neuroimaging and neuroanatomy have yielded major insights concerning fundamental principles of cortical organization and evolution, thus speaking to how well different species serve as models for human brain function in health and disease. Here, we focus on cortical folding, parcellation, and connectivity in mice, marmosets, macaques, and humans. Cortical folding patterns vary dramatically across species, and individual variability in cortical folding increases with cortical surface area. Such issues are best analyzed using surface-based approaches that respect the topology of the cortical sheet. Many aspects of cortical organization can be revealed using 1 type of information (modality) at a time, such as maps of cortical myelin content. However, accurate delineation of the entire mosaic of cortical areas requires a multimodal approach using information about function, architecture, connectivity, and topographic organization. Comparisons across the 4 aforementioned species reveal dramatic differences in the total number and arrangement of cortical areas, particularly between rodents and primates. Hemispheric variability and bilateral asymmetry are most pronounced in humans, which we evaluated using a high-quality multimodal parcellation of hundreds of individuals. Asymmetries include modest differences in areal size but not in areal identity. Analyses of cortical connectivity using anatomical tracers reveal highly distributed connectivity and a wide range of connection weights in monkeys and mice; indirect measures using functional MRI suggest a similar pattern in humans. Altogether, a multifaceted but integrated approach to exploring cortical organization in primate and nonprimate species provides complementary advantages and perspectives.
PUBLICATION:
Proceedings of the National Academy of Sciences of the United States of America
- DOI:
10.1073/pnas.1902299116
- PMID:
31871175
- David C Van Essen
- Chad J Donahue
- Timothy S Coalson
- Henry Kennedy
- Takuya Hayashi
- Matthew F Glasser
- RIKEN
- University of Lyon
- Institute of Neuroscience, Shanghai
- Washington University in St. Louis
-
VanEssen_MonkeyModels_PNAS2019.scene
SCENES: -
VanEssen_MonkeyModels_PNAS2019_SI.scene
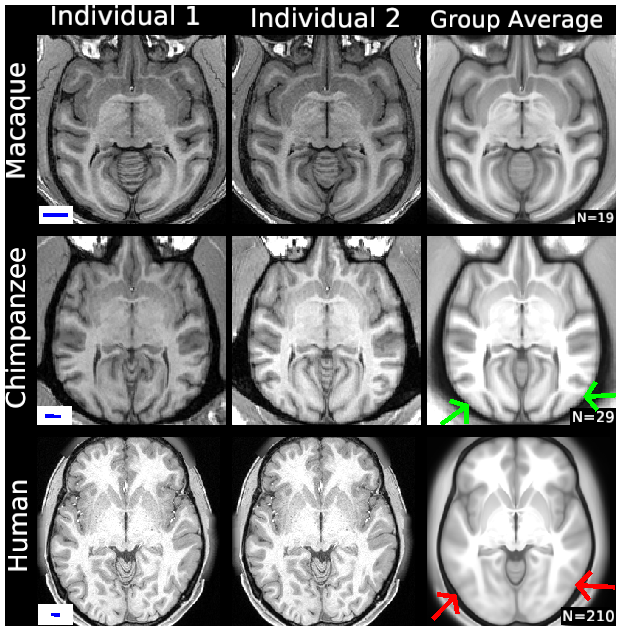
SCENES:- Figure S1. Individual variability in structural MRI volume slices
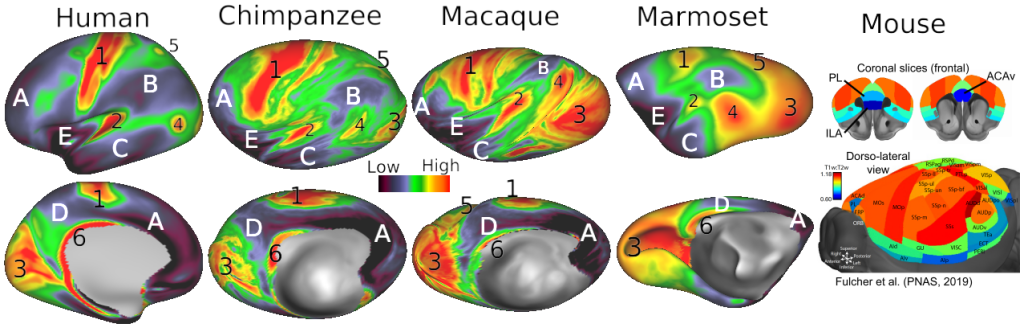
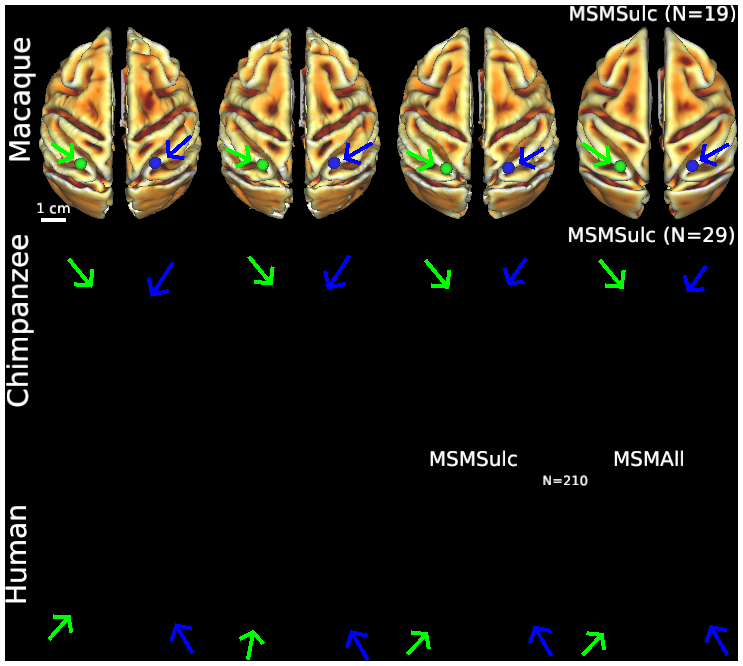
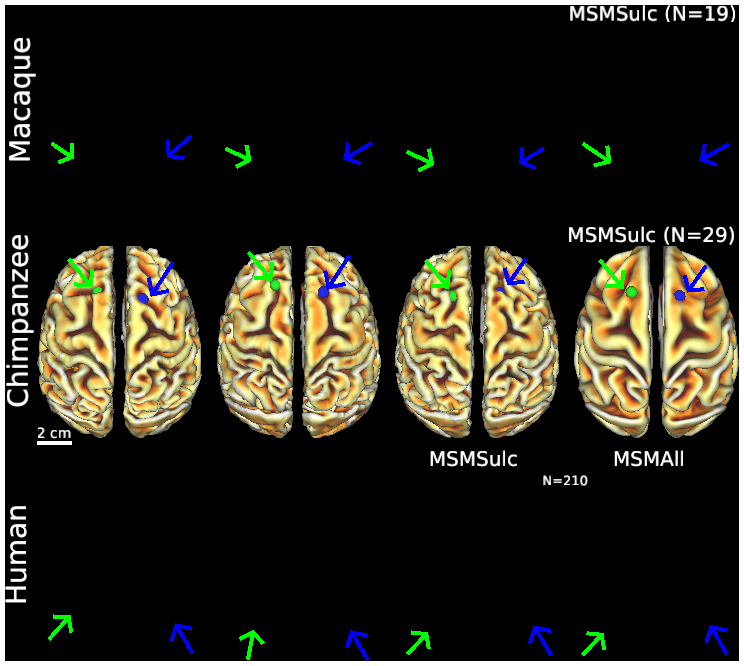
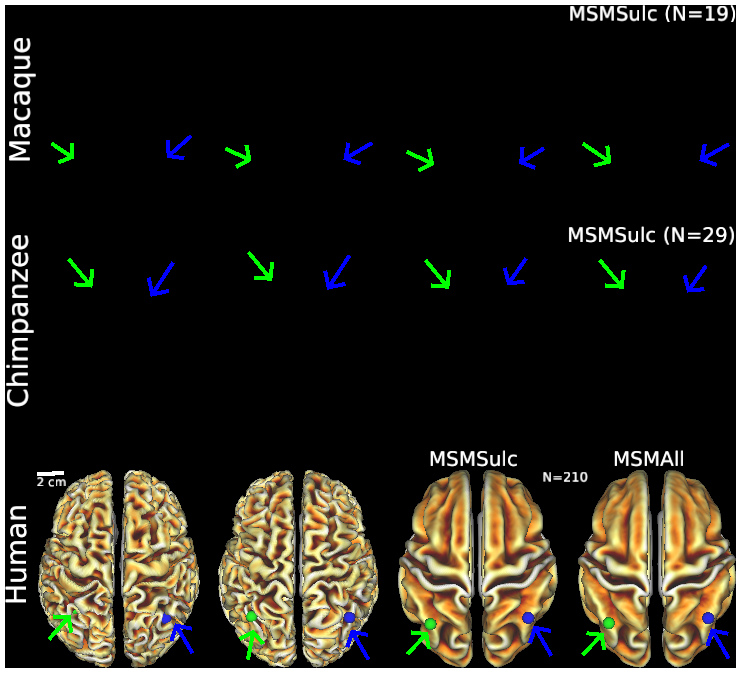
- Figure S2 (top row) Macaque folding fariability - surface views
- Figure S2 (middle row) Chimpanzee folding variability - surface views
- Figure S2 (bottom row) Human folding variability - surface views
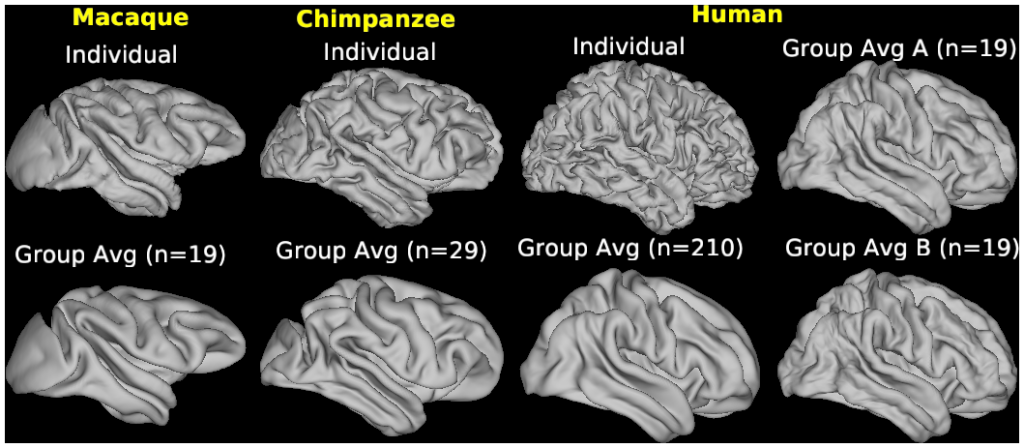
- Figure S3 Effects of sample size on human group average surfaces