study:
Cerebral Cortical Folding, Parcellation, and Connectivity
SCENE FILE:
VanEssen_MonkeyModels_PNAS2019_SI
SCENE:
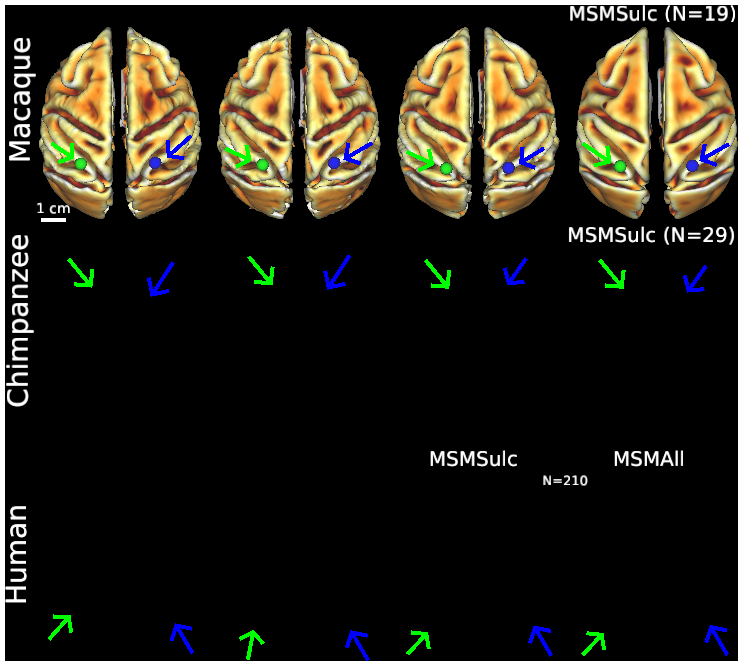
Figure S2 (top row) Macaque folding fariability - surface views
DESCRIPTION:
Individual variability of cortical folding shown on dorsal views of cortical midthickness surface reconstructions and surface-based registration to an atlas, using MSMSulc folding-based registration for all 3 species (Donahue et al., PNAS, 2018; Glasser et al., Nature, 2016) and also MSMAll areal feature based registration for humans (Van Essen et al., Cerebral Cortex, 2012; Robinson et al., Neuroimage, 2018). Scale bars (2 cm) on left apply to all individuals in that row. In the top row, the left and right midthickness surfaces for the three individual macaques are highly similar in shape except for local irregularities reflecting minor segmentation artifacts. The Yerkes19 group-average left and right midthickness surfaces (top right, aligned using folding features) are slightly smoother but otherwise nearly identical to the individuals. The strong bilateral symmetry in the group average surfaces was used to generate standard-mesh representations that have excellent left-right geographic correspondence (Van Essen et al., Cerebral Cortex, 2012) using GIFTI and CIFTI data formats (Glasser et al., Neuroimage, 2013). For example, the blue vertex identified on the right group average inferior parietal lobule lies in precisely corresponding locations in all three individual right hemispheres (blue vertices, arrows) and in closely corresponding locations in the left hemisphere atlas and individuals (green vertices, arrows).
TAGS:
Surface Mesh:32k fs LR, Species:Chimpanzee, Species:Human, Registration:MSMAll, Species:Macaque, Atlas:Yerkes 19, Registration:MSMSulc, Other Data:gene
