FULL TITLE:
The NonHuman Primate Neuroimaging & Neuroanatomy Project
SPECIES:
Human, Marmoset, Chimpanzee, Macaque
DESCRIPTION:
The scenes for this study were created using a newer version of wb_view, they will not display properly using wb_view v1.4.2. Please keep an eye out for the new wb_view release coming early 2021.
ABSTRACT:
Multi-modal neuroimaging projects such as the Human Connectome Project (HCP) and UK Biobank are advancing our understanding of human brain architecture, function, connectivity, and their variability across individuals using high-quality non-invasive data from many subjects. Such efforts depend upon the accuracy of non-invasive brain imaging measures. However, 'ground truth' validation of connectivity using invasive tracers is not feasible in humans. Studies using nonhuman primates (NHPs) enable comparisons between invasive and non-invasive measures, including exploration of how "functional connectivity" from fMRI and "tractographic connectivity" from diffusion MRI compare with long-distance connections measured using tract tracing. Our NonHuman Primate Neuroimaging & Neuroanatomy Project (NHP_NNP) is an international effort (6 laboratories in 5 countries) to: (i) acquire and analyze high-quality multi-modal brain imaging data of macaque and marmoset monkeys using protocols and methods adapted from the HCP; (ii) acquire quantitative invasive tract-tracing data for cortical and subcortical projections to cortical areas; and (iii) map the distributions of different brain cell types with immunocytochemical stains to better define brain areal boundaries. We are acquiring high-resolution structural, functional, and diffusion MRI data together with behavioral measures from over 100 individual macaques and marmosets in order to generate non-invasive measures of brain architecture such as myelin and cortical thickness maps, as well as functional and diffusion tractography-based connectomes. We are using classical and next-generation anatomical tracers to generate quantitative connectivity maps based on brain-wide counting of labeled cortical and subcortical neurons, providing ground truth measures of connectivity. Advanced statistical modeling techniques address the consistency of both kinds of data across individuals, allowing comparison of tracer-based and non-invasive MRI-based connectivity measures. We aim to develop improved cortical and subcortical areal atlases by combining histological and imaging methods. Finally, we are collecting genetic and sociality-associated behavioral data in all animals in an effort to understand how genetic variation shapes the connectome and behavior.
PUBLICATION:
NeuroImage
- DOI:
10.1016/j.neuroimage.2021.117726
- PMID:
33484849
- Takuya Hayashi
- Yujie Hou
- Matthew F Glasser
- Joonas A Autio
- Kenneth Knoblauch
- Miho Inoue-Murayama
- Tim Coalson
- Essa Yacoub
- Stephen Smith
- Henry Kennedy
- David C Van Essen
- University of Minnesota
- Institute of Neuroscience, Shanghai
- Washington University in St. Louis
- University of Lyon
- University of Oxford
- Kyoto University
- RIKEN
-
Figures.scene
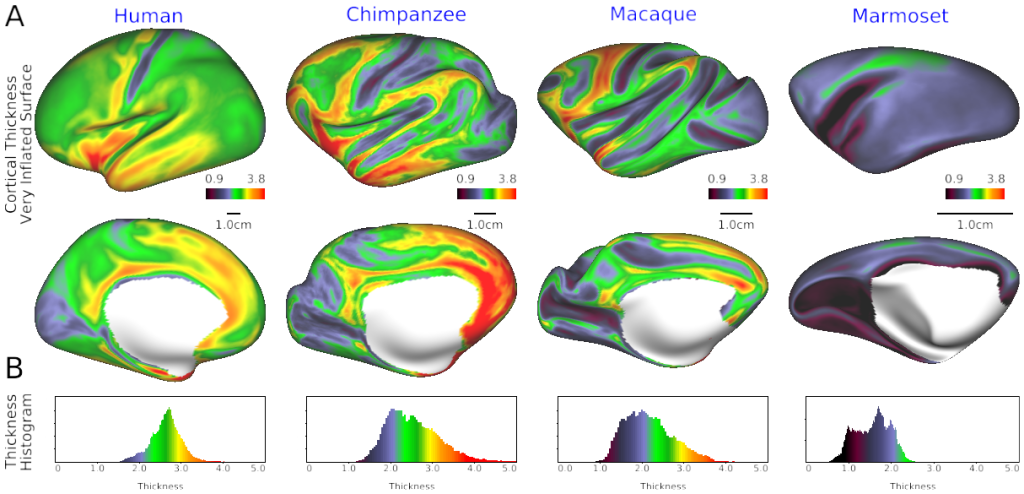
SCENES:- Fig. 1 Cortical thickness in human, chimpanzee, macaque and marmoset
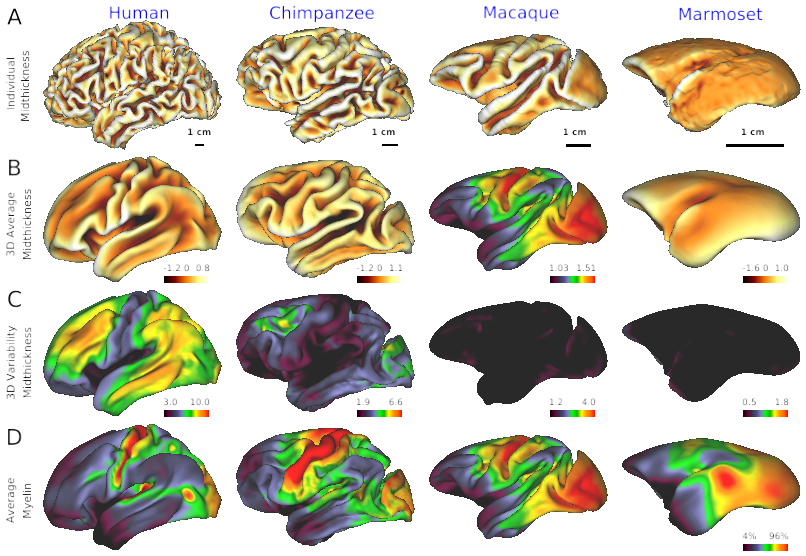
- Fig. 2 Cross-species comparison of individual’s cortical surface, average and variability
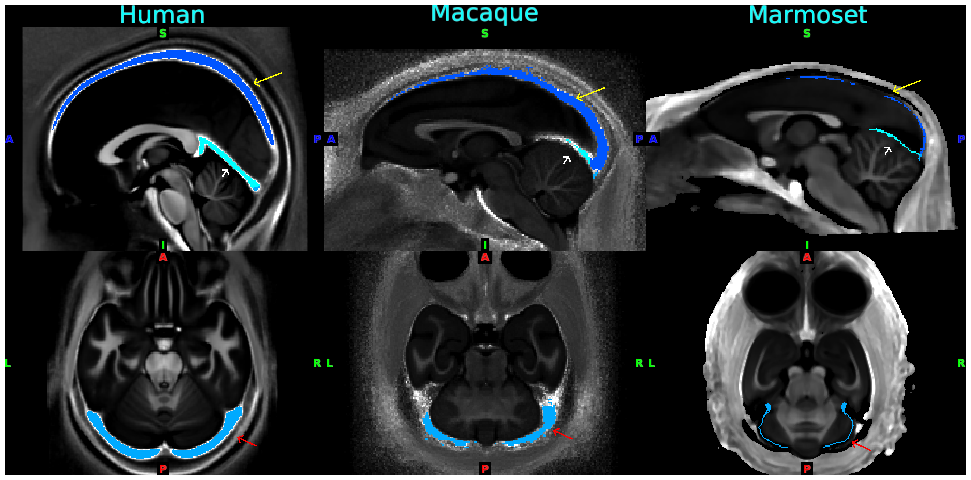
- Fig. 3 Standardized venous sinus maps for each species of human, macaque and marmoset
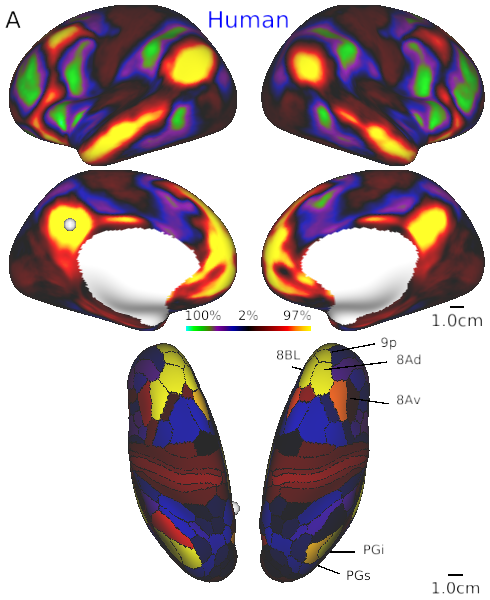
- Fig. 4A Default mode network in human
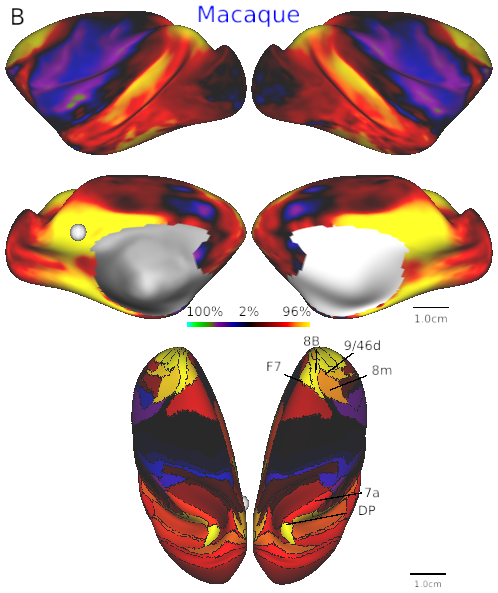
- Fig. 4B Default mode network in macaque.
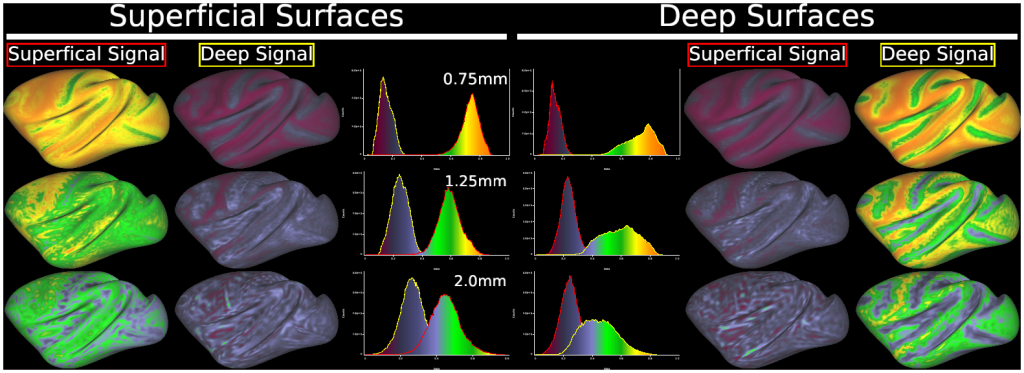
- Fig. 8 Results of simulation for partial volume/geometric effects in ‘layer’ fMRI analysis at various resolutions in macaque cerebral cortex.