FULL TITLE:
Connectome Workbench v1.5 Tutorial
SPECIES:
Human
DESCRIPTION:
This dataset contains HCP-generated data for completing the Connectome Workbench v1.5 Tutorial. A PDF of the tutorial instructions and data Release notes are also included. Scenes 5 & 6 for viewing functional connectivity (see below) require the user to have internet access and a ConnectomeDB account. Sign up for ConnectomeDB at https://db.humanconnectome.org/
Data included from the HCP-Young Adult 500 Subjects Release:
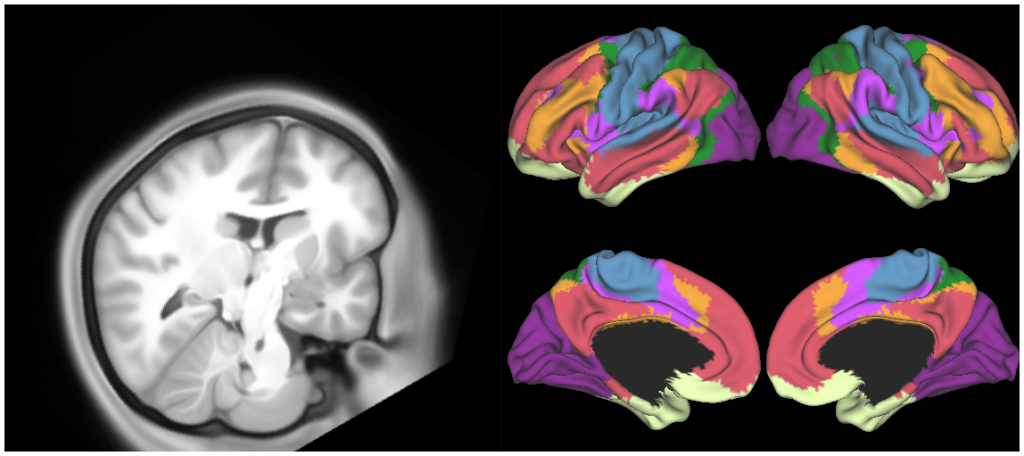
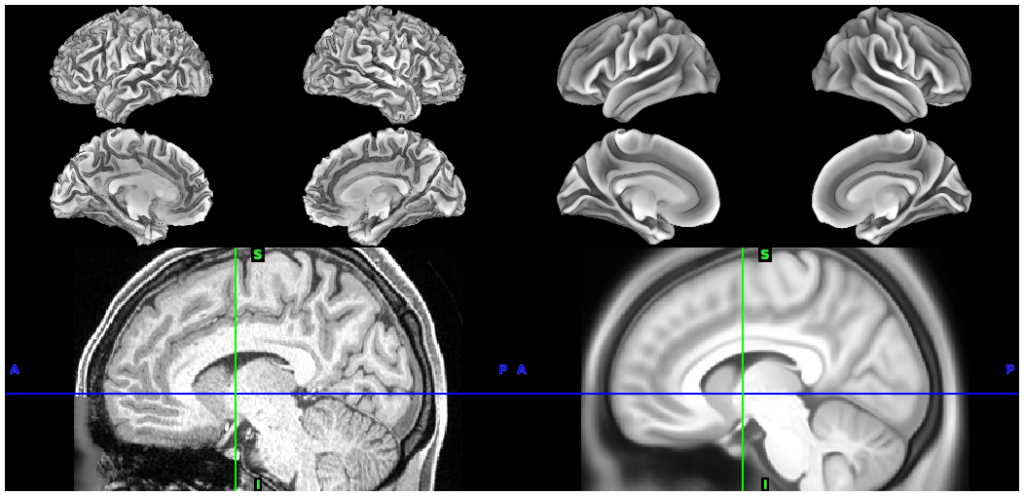
*individual subject (100307) and 440 subject (R440) group-average structural files (including surfaces at 32k and 164k mesh resolutions)
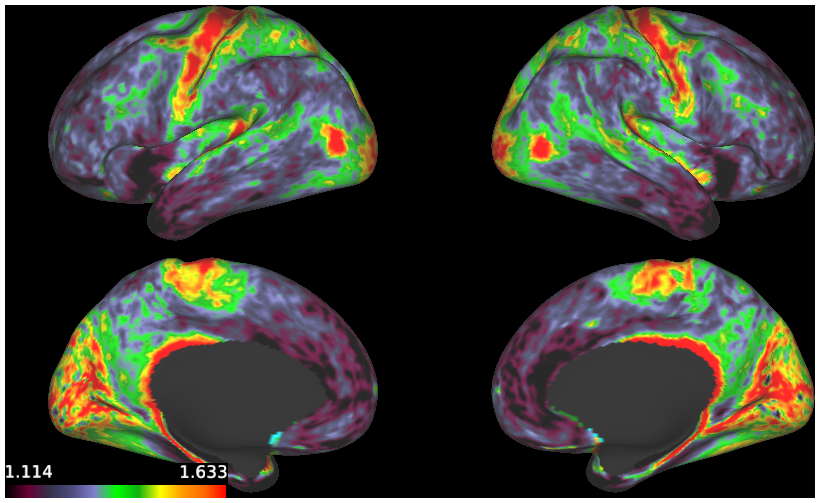
*maps of myelin, curvature, Freesurfer sulc and cortical thickness for 100307 and R440 (at 32k and 164k mesh resolutions)
*groupaverage tfMRI grayordinates analysis results for R440
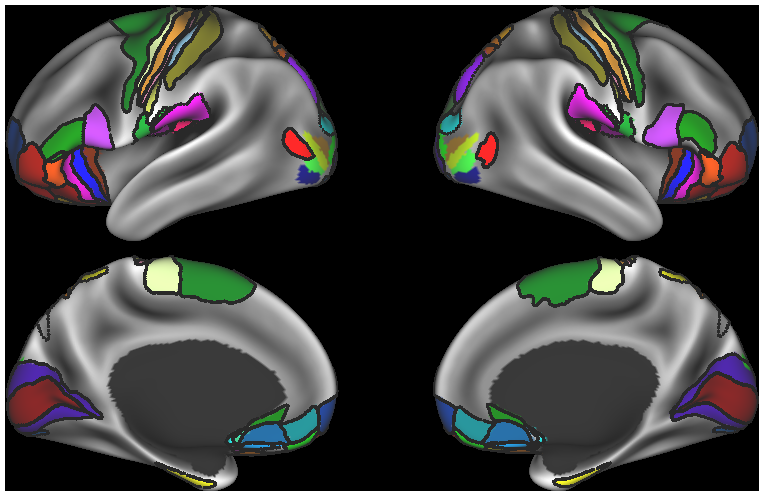
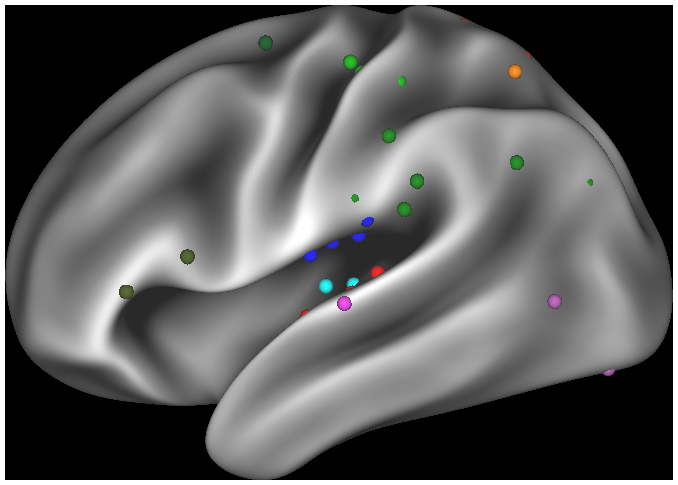
*label, borders, and foci files generated from other studies to use for reference
Due to their large size (33 GB each), the two group-average dense functional connectome files for R468 are remotely accessed in ConnectomeDB (shown in Scenes 5 & 6) to generate connectivity maps ‘on the fly’, requiring the user to have internet access and login using their ConnectomeDB account credentials.
For those that would like a local copy of similar S1200 versions of the dense connectome files or the Group-PCA Eigenmaps (2 GB) that can be used to create them locally using wb_command, they are available for download from the WU-Minn HCP Data -- 1200 Subjects project page in ConnectomeDB: https://db.humanconnectome.org/data/projects/HCP_1200
- John Harwell
- Jennifer Elam
- David Van Essen
- Erin Reid
- Timothy Coalson
- Matthew Glasser
- Washington University in St. Louis
-
WB_1.5_SCENES.32k_fs_LR.scene
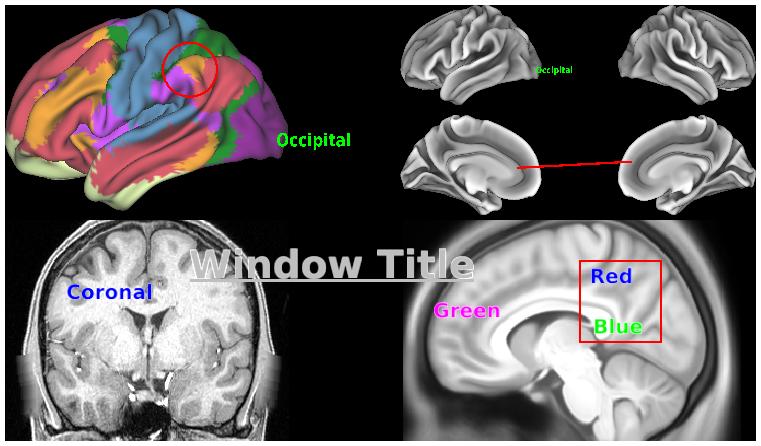
SCENES:- 1. Individual surfaces and volumes
- 2. Individual vs Group Avg surfaces and volumes
- 3. Individual vs Group Avg Myelin Maps on R440 Surfaces
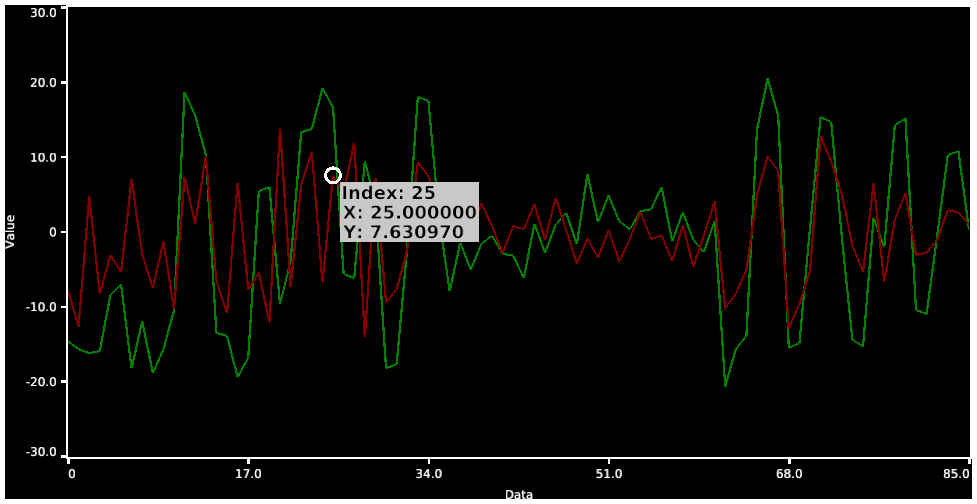
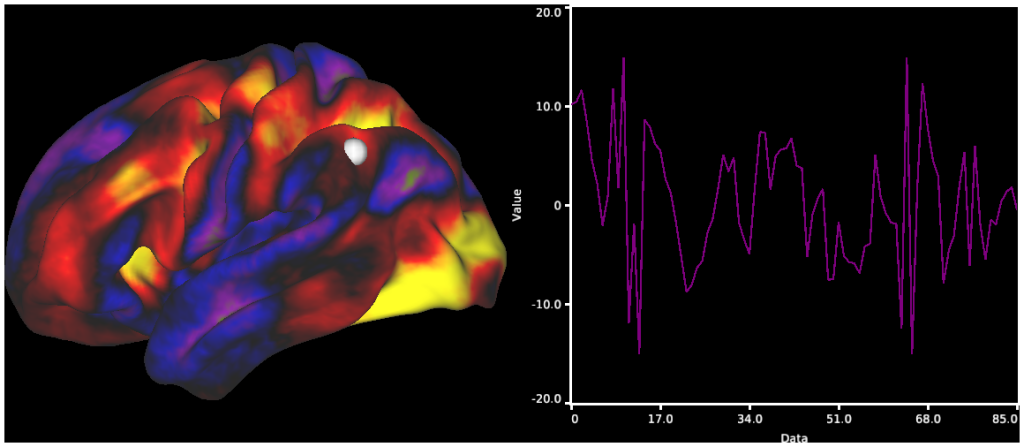
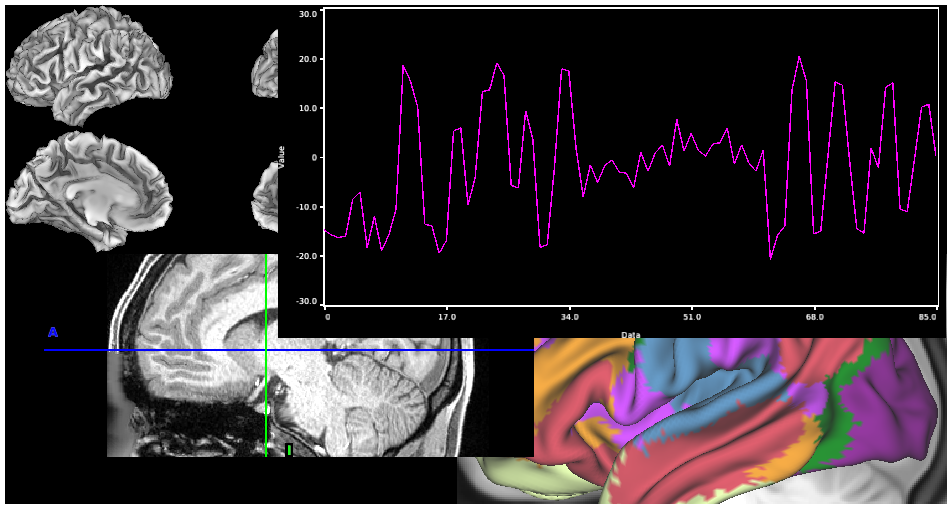
- 4. Resting State fMRI Time Series surfaces, volume, and charting
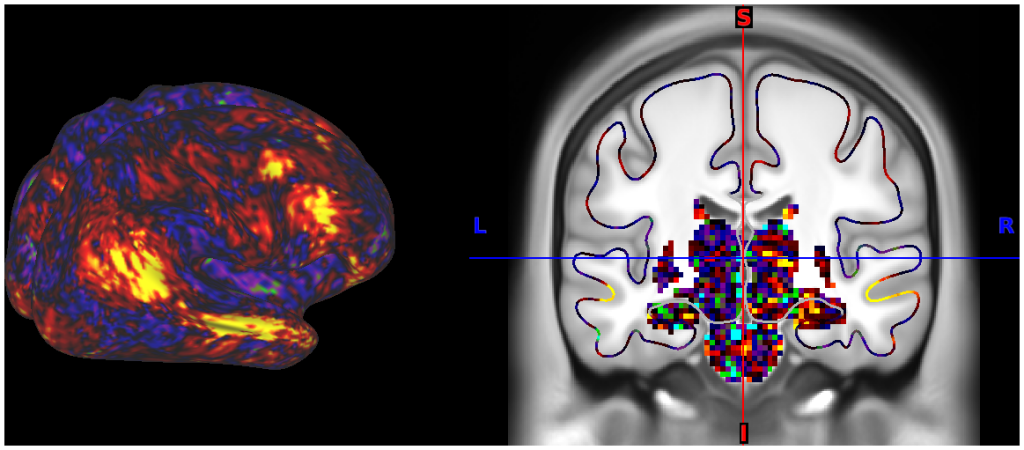
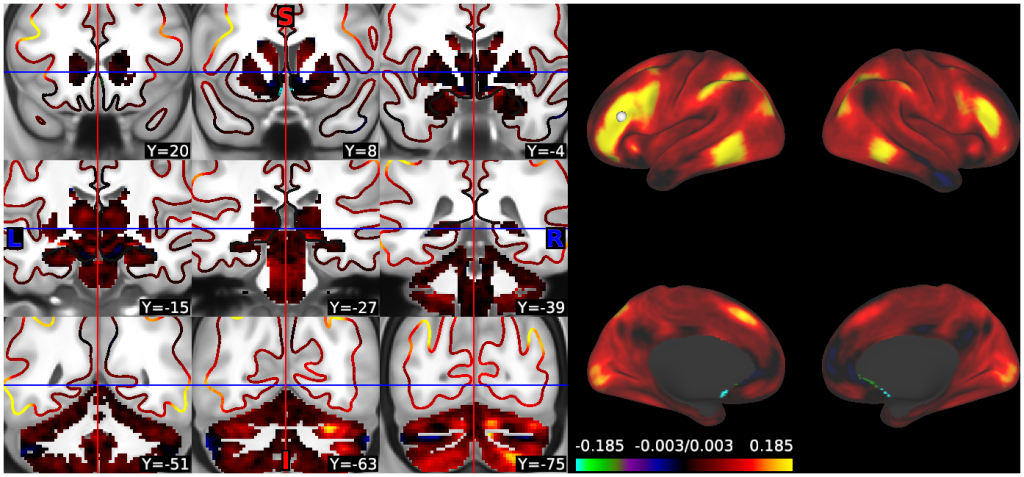
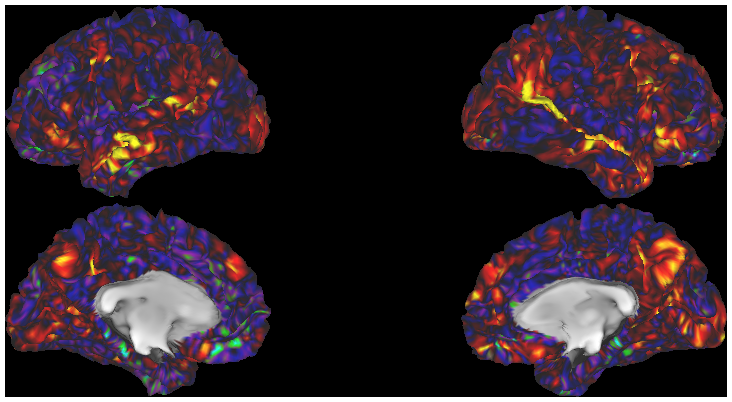
- 5. fcMRI, Full Correlation, R468 surfaces and volume montage
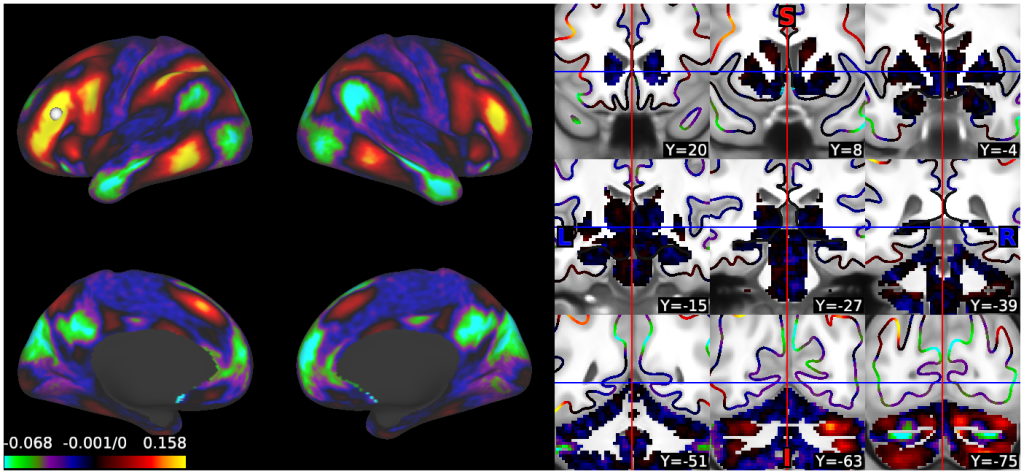
- 6. fcMRI, MGT-regressed correlation, R468 surfaces and volume montage
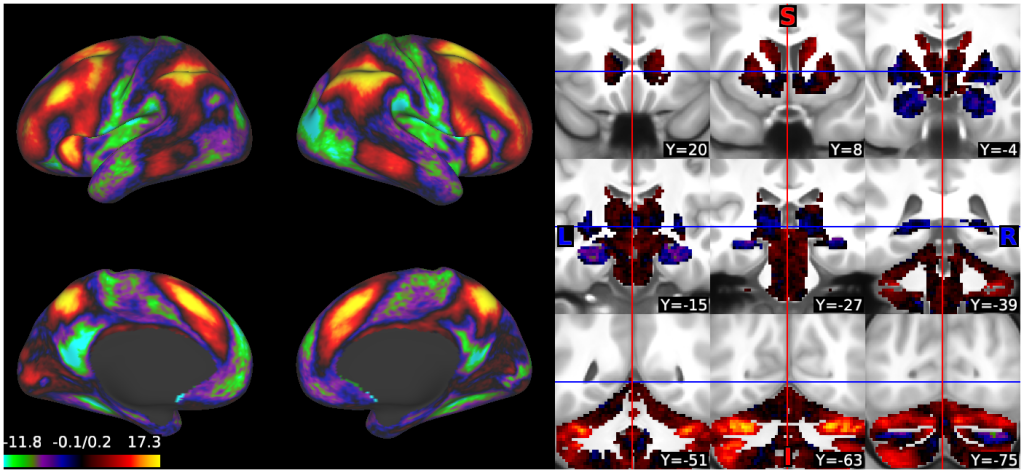
- 7. tfMRI (2BK-0BK WM), R440, on surfaces and volume montage
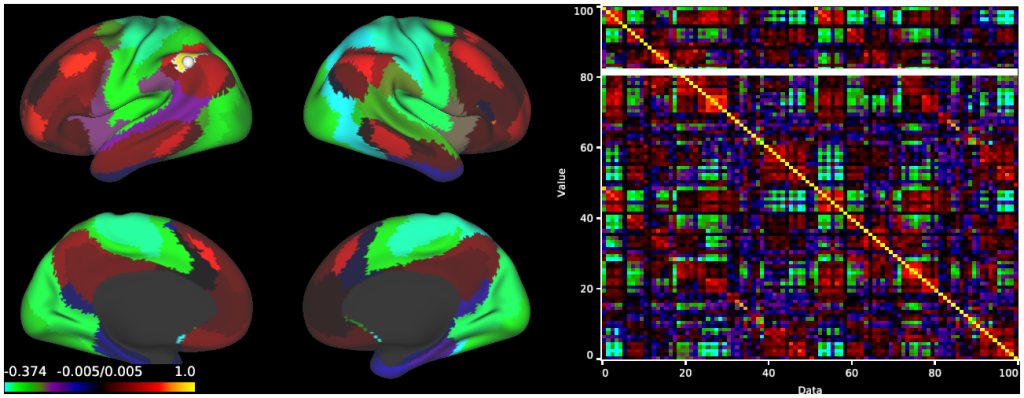
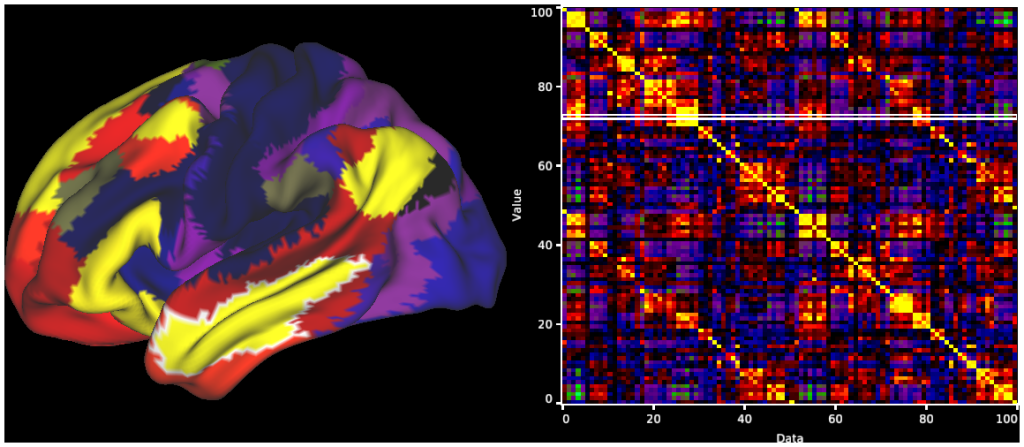
- 8. Parcellated MGTR Connectivity, R196, RSN parcels: Surface and Matrix chart display
- 9. Parcellation Borders displayed with parcellation labels on R440 surfaces
- 10. Foci displayed on R440 surfaces
- Annotations
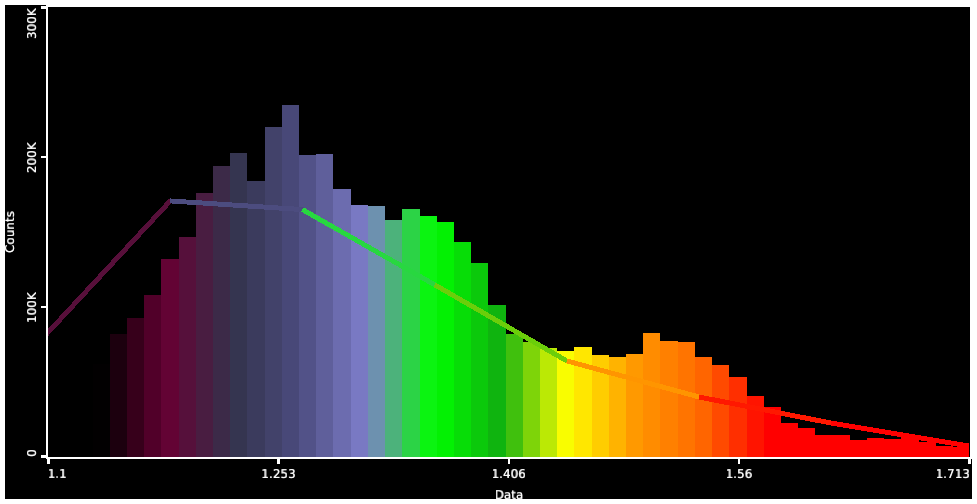
- Histogram Chart
- Lines Chart
- Dynamic Lines Chart
- Matrix Chart
- Media (Image) in Left tab
- Manual Tile Tabs Layout
- Dynamic Connectivity
-
WB_1.5_SCENES.164k_fs_LR.scene
SCENES: