FULL TITLE:
Human HCP 1200 Group Average + Individuals; Structural + fMRI Atlas
SPECIES:
Human
DESCRIPTION:
This Human reference dataset comprises data from the Human Connectome Project (HCP) 1200 Subjects (S1200) data release (March 2017) processed using HCP pipelines. It includes:
1) Group-average structural and functional (task) MRI data (HCP_S1200_GroupAvg_BALSA.scene, ID: D4w0w)
2) Selected individual-subject anatomical maps for each of the 1096 subjects having at least 1 rfMRI run (HCP_S1200_Individuals_BALSA.scene, ID: kNzvk)
Given the large differences in file size, downloading only the first Group-average scene file and its associated data files is recommended unless the individual-subject data files are specifically needed.
The S1200 dataset is the complete HCP young adult data release and includes imaging data registered using both 1) cortical areal feature-based surface registration (“MSMAll”, using folding, myelin maps, and rfMRI), and 2) purely folding-based registration (“MSMSulc”) used for the S500 and earlier HCP data releases; MSM stands for Multimodal Surface Matching (Robinson et al., Neuroimage, 2014, 2018). Results from MSMSulc and MSMAll may be compared in the task-fMRI scene in HCP_S1200_GroupAvg_BALSA.scene
Modestly different numbers of HCP subjects were used for the analysis of the different modalities. Group average and individual structural data are available on 1096 subjects that have at least one run of rfMRI data (and hence were eligible for MSMAll registration). Task fMRI data is based on 997 subjects that additionally have complete tfMRI data for all 7 HCP tasks. Since recruitment for the HCP was focused on families with at least one twin pair born in Missouri, USA, these groups contain some subjects that are related to some members of the group, thus they are referred to as R1096 and R997 (‘R’ designating related).
Individual S1200 subjects structural imaging data in HCP_S1200_Individuals_BALSA.scene may be scrolled through in wb_view efficiently as separate maps within each of several S1200.All.*.dscalar.nii files.
Detailed information on collection, processing, and analysis of this data is available in the HCP 1200 Subjects Release Reference Manual at: https://www.humanconnectome.org/storage/app/media/documentation/s1200/HCP_S1200_Release_Reference_Manual.pdf
HCP pipelines code available at: https://github.com/Washington-University/HCPpipelines
Key references:
Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR, Van Essen DC, Jenkinson M; WU-Minn HCP Consortium. (2013) The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage 80:105-24. (doi: 10.1016/j.neuroimage.2013.04.127) PMID: 23668970
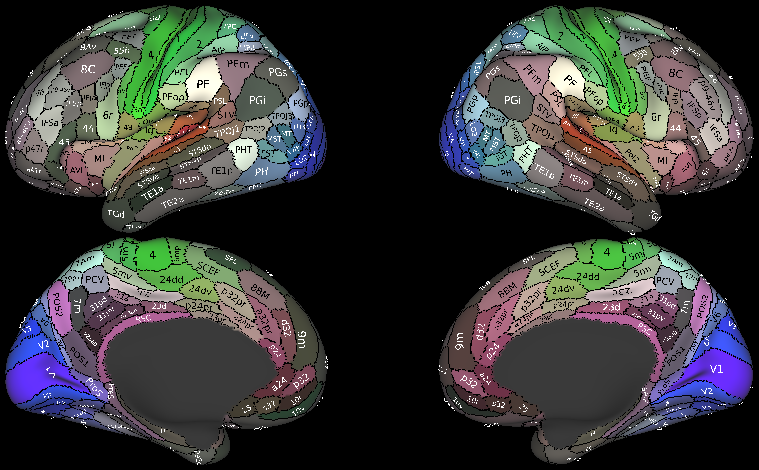
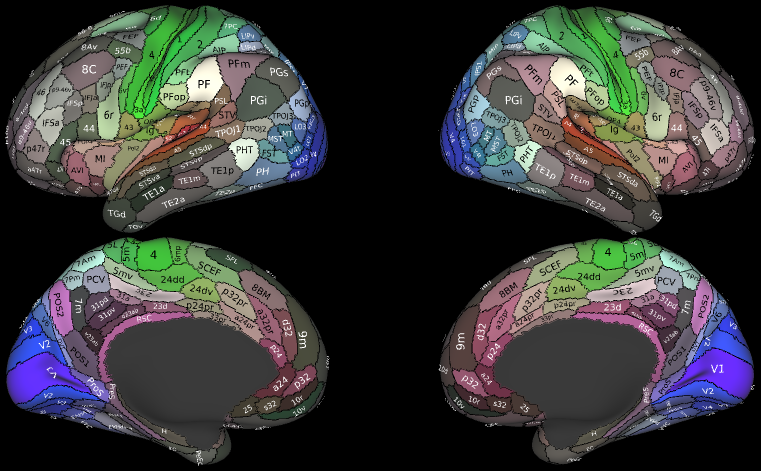
Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, Ugurbil K, Andersson J, Beckmann CF, Jenkinson M, Smith SM, Van Essen DC. (2016) A multi-modal parcellation of human cerebral cortex. Nature. 536(7615):171-178. (doi: 10.1038/nature18933) PMID: 27437579
Gordon EM, Laumann TO, Adeyemo B, Huckins JF, Kelley WM, Petersen SE (2016) Generation and Evaluation of a Cortical Area Parcellation from Resting-State Correlations. Cereb Cortex.26:288-303. (doi: 10.1093/cercor/bhu239) PMID: 25316338
Ji JL, Spronk M, Kulkarni K, Repovš G, Anticevic A, Cole MW (2019) Mapping the human brain's cortical-subcortical functional network organization. Neuroimage 185:35-57. (doi: 10.1016/j.neuroimage.2018.10.006) PMID: 30291974
Robinson EC, Jbabdi S, Glasser MF, Andersson J, Burgess GC, Harms MP, Smith SM, Van Essen DC, Jenkinson M. (2014) MSM: a new flexible framework for Multimodal Surface Matching. Neuroimage 100:414-26. (doi: 10.1016/j.neuroimage.2014.05.069) PMID: 24939340
Van Essen, DC, Glasser, MF, Dierker, D, Harwell, J, and Coalson, T (2012) Parcellations and hemispheric asymmetries of human cerebral cortex analyzed on surface-based atlases. Cerebral Cortex 22: 2241-2262. (doi: 10.1093/cercor/bhr291) PMID: 22062192
Yeo et al. (2011): Yeo BTT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zöllei L, Polimeni JR et al. 2011. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 106: 1125–1165.
-
HCP_S1200_GroupAvg_BALSA.scene
DESCRIPTION: Human Surface Atlas + Group average Anatomical maps, tfMRI maps
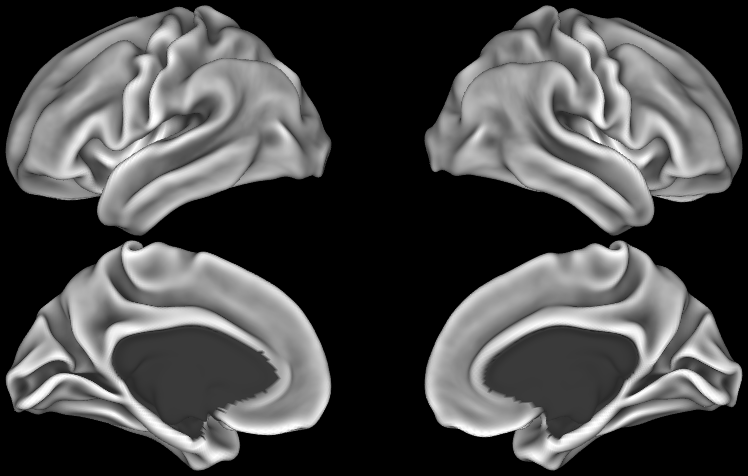
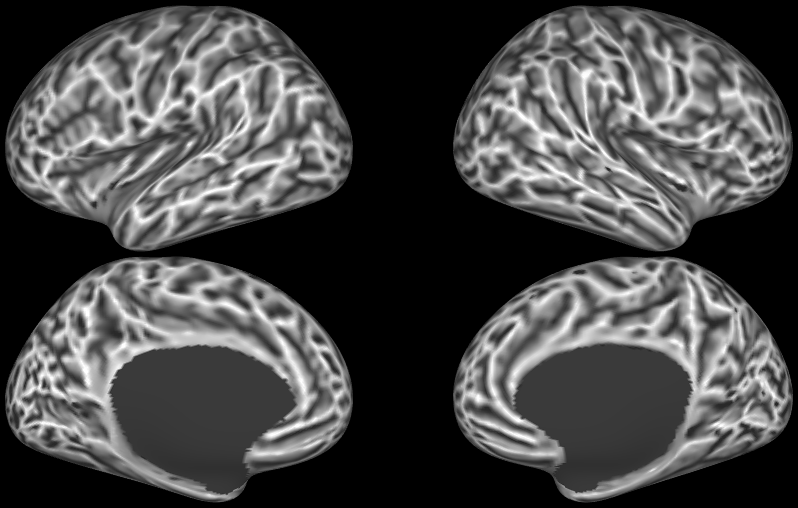
SCENES:- HCP S1200 Average midthickness surfaces, folding overlay + average volumes (1096 subjects, MSM-All Registered)
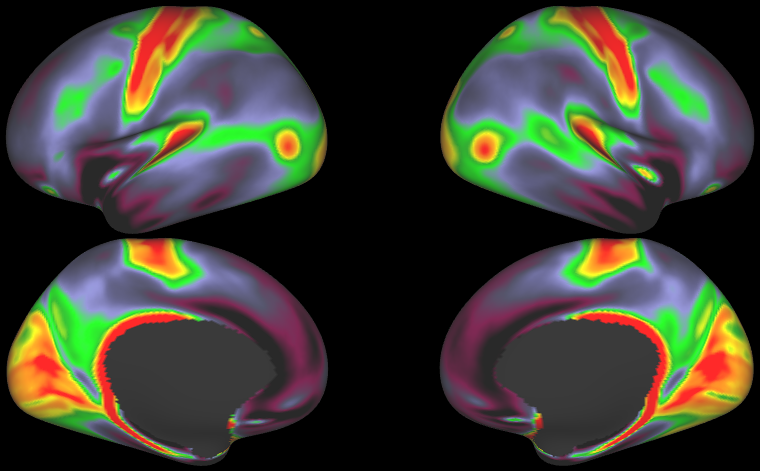
- HCP S1200 Group Average Maps: myelin, thickness, curvature, sulc (1096 subjects, MSM-All Registered)
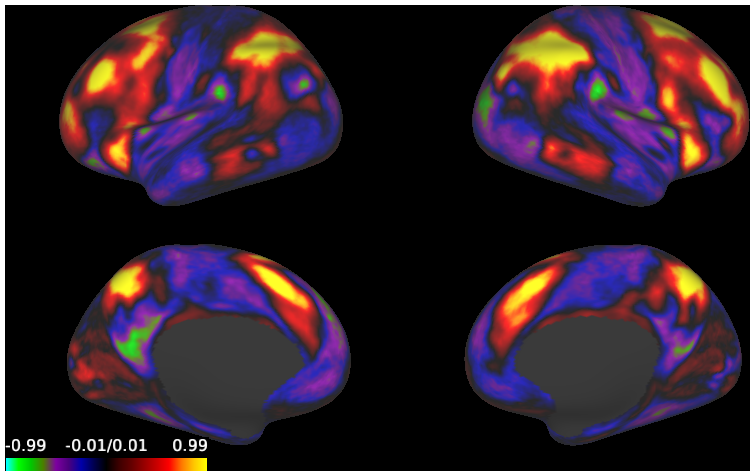
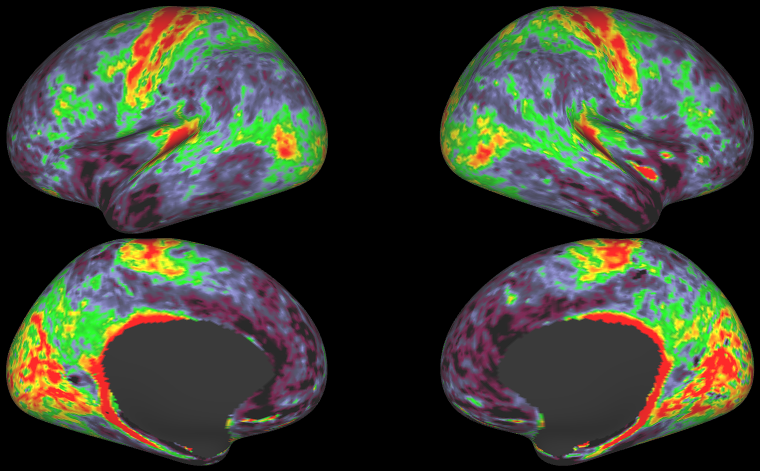
- HCP S1200 Average task-fMRI Cohen's D effect-size maps (997 subjects, MSMAll vs. MSMSulc Registered)
- Cortical Parcellations
-
HCP_S1200_Individuals_BALSA.scene
DESCRIPTION: Human HCP S1200 Individual Anatomical maps
SCENES: