FULL TITLE:
Parcellating Cerebral Cortex: How Invasive Animal Studies Inform Non-Invasive Map-making in Humans
SPECIES:
Human
ABSTRACT:
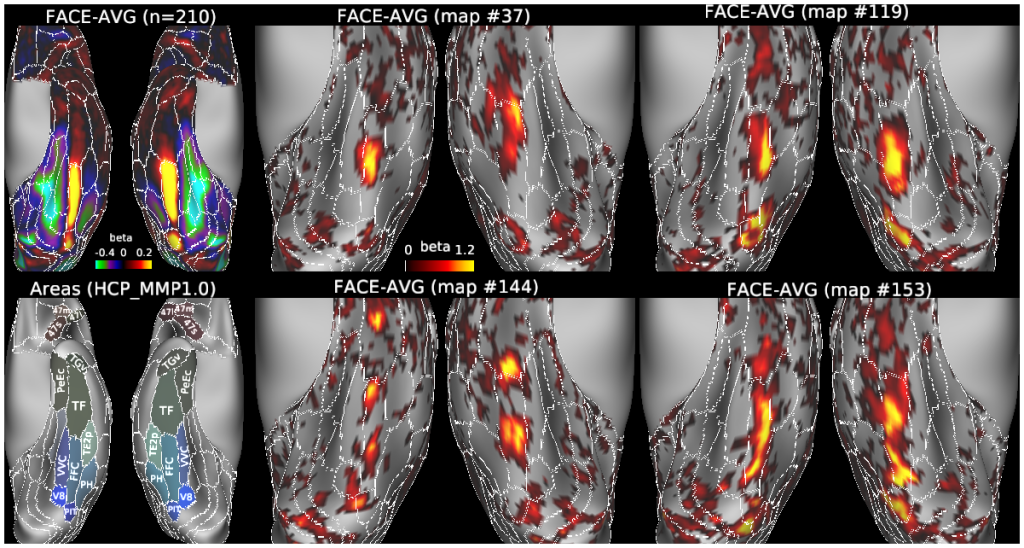
Cerebral cortex in mammals contains a mosaic of cortical areas that differ in function, architecture, connectivity, and/or topographic organization. A combination of local connectivity (within-area microcircuitry) and long-distance (between-area) connectivity enables each area to perform a unique set of computations. Some areas also have characteristic within-area mesoscale organization, reflecting specialized representations of distinct types of information. Cortical areas interact with one another to form functional networks that mediate behavior, and each area may be a part of multiple, partially overlapping networks. Given their importance to understanding brain organization, mapping cortical areas across species is a major objective of systems neuroscience and has been a century-long challenge. Here we review recent progress in multi-modal mapping of mouse and nonhuman primate cortex, mainly using invasive experimental methods. These studies also provide a neuroanatomical foundation for mapping human cerebral cortex using non-invasive neuroimaging, including a new map of human cortical areas that we generated using a semi-automated analysis of high quality multi-modal neuroimaging data. We contrast our semi-automated approach to human multi-modal cortical mapping with various extant fully automated human brain parcellations that are based on only a single imaging modality, and offer suggestions on how to best advance the non-invasive brain parcellation field. We discuss the limitations as well as strengths of current non-invasive methods of mapping brain function, architecture, connectivity, and topography and of current approaches to mapping the brain’s functional networks.
PUBLICATION:
Neuron
- David C Van Essen
- Matthew F Glasser
- Washington University in St. Louis