FULL TITLE:
Multimodal 3D atlas of the macaque monkey motor and premotor cortex
SPECIES:
Macaque
DESCRIPTION:
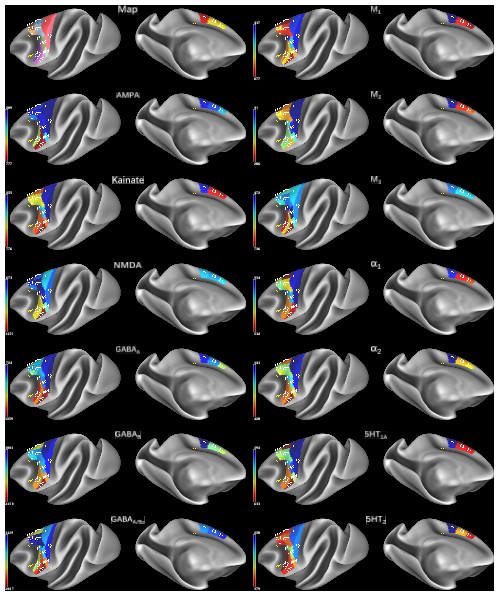
Parcellation scheme of the macaque motor cortex projected onto the lateral and medial views of the Yerkes19 surface (Donahue et al., 2016). 16 cyto- and receptor- architectonically distinct areas were identified in this brain region: areas 4a, 4p and 4m of the primary motor cortex), a supplementary motor area (F3), a pre-supplementary motor area (F6), dorsolateral premotor areas F2d, F2v, F7d, F7i, and F7s, as well as ventrolateral premotor areas F4s, F4d, F4v, F5s, F5d, and F5v). The mean receptor densities of receptors for glutamate (AMPA, kainate, NMDA), GABA (GABAA, GABAB, GABAA associated benzodiazepine (GABAA/BZ) binding sites), acetylcholine (M1, M2, M3), norepinephrine (alpha1, alpha2) and serotonin (5-HT1A, 5-HT2) have been projected onto the corresponding area. Color bars code for receptor densities in fmol/mg protein.
cgs - cingulate sulcus, ias - inferior arcuate branch, lf - lateral fissure, sas - superior arcuate branch, sts - superior temporal sulcus.
To view larger versions of each receptor density map, select View>Exit Tile Tabs and click on tabs to navigate to desired map. See the second layer in the Overlay Toolbox for the name of the map file displayed.
ABSTRACT:
In the present study, we reevaluated the parcellation scheme of the macaque frontal agranular cortex by implementing quantitative cytoarchitectonic and multireceptor analyses, with the purpose to integrate and reconcile the discrepancies between previously published maps of this region. We applied an observer-independent and statistically testable approach to determine the position of cytoarchitectonic borders. Analysis of the regional and laminar distribution patterns of 13 different transmitter receptors confirmed the position of cytoarchitectonically identified borders. Receptor densities were extracted from each area and visualized as its "receptor fingerprint". Hierarchical and principal components analyses were conducted to detect clusters of areas according to the degree of (dis)similarity of their fingerprints. Finally, functional connectivity pattern of each identified area was analyzed with areas of prefrontal, cingulate, somatosensory and lateral parietal cortex and the results were depicted as "connectivity fingerprints" and seed-to-vertex connectivity maps. We identified 16 cyto- and receptor-architectonically distinct areas, including novel subdivisions of the primary motor area 4 (i.e. 4a, 4p, 4m) and of premotor areas F4 (i.e. F4s, F4d, F4v), F5 (i.e. F5s, F5d, F5v) and F7 (i.e. F7d, F7i, F7s). Multivariate analyses of receptor fingerprints revealed three clusters, which first segregated the subdivisions of area 4 with F4d and F4s from the remaining premotor areas, then separated ventrolateral from dorsolateral and medial premotor areas. The functional connectivity analysis revealed that medial and dorsolateral premotor and motor areas show stronger functional connectivity with areas involved in visual processing, whereas 4p and ventrolateral premotor areas presented a stronger functional connectivity with areas involved in somatomotor responses. For the first time, we provide a 3D atlas integrating cyto- and multi-receptor-architectonic features of the macaque motor and premotor cortex. This atlas constitutes a valuable resource for the analysis of functional experiments carried out with non-human primates, for modeling approaches with realistic synaptic dynamics, as well as to provide insights into how brain functions have developed by changes in the underlying microstructure and encoding strategies during evolution.
PUBLICATION:
NeuroImage
- DOI:
10.1016/j.neuroimage.2020.117574
- PMID:
33221453
- Lucija Rapan
- Sean Froudist-Walsh
- Meiqi Niu
- Ting Xu
- Thomas Funck
- Karl Zilles
- Nicola Palomero-Gallagher
- New York University
- Heinrich-Heine University
- Research Centre Jülich
- Child Mind Institute
- Medical Faculty, RWTH Aachen