FULL TITLE:
Hierarchy of transcriptomic specialization across human cortex captured by structural neuroimaging topography
SPECIES:
Human, Macaque
DESCRIPTION:
The scenes included in this study illustrate the cortical parcellations, monkey and human structural neuroimaging maps, tractography data, and pre-processed gene expression profiles which were used in the primary analyses reported in Burt et al. (2018) Nat Neurosci. A brief description of each scene file is provided below:
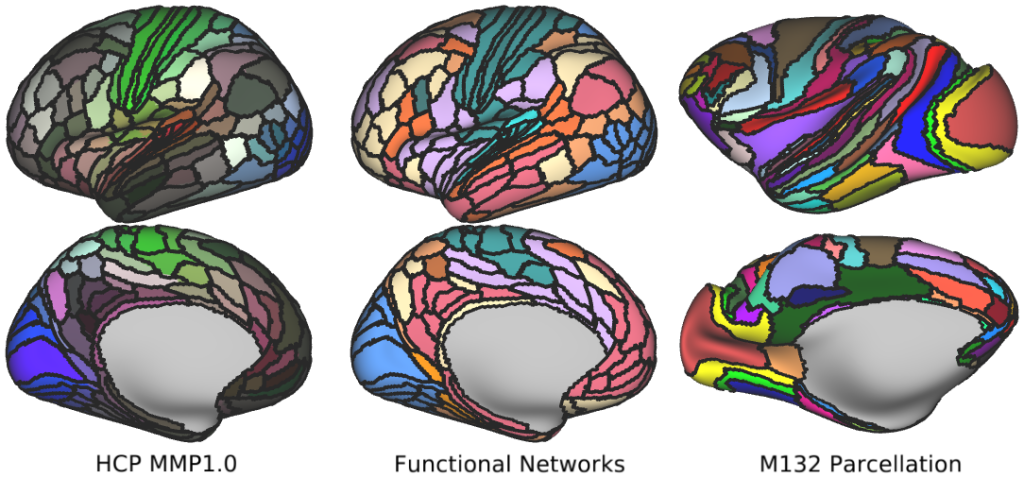
Cortical Parcellations -- Human and monkey cortical parcellations used throughout the text.
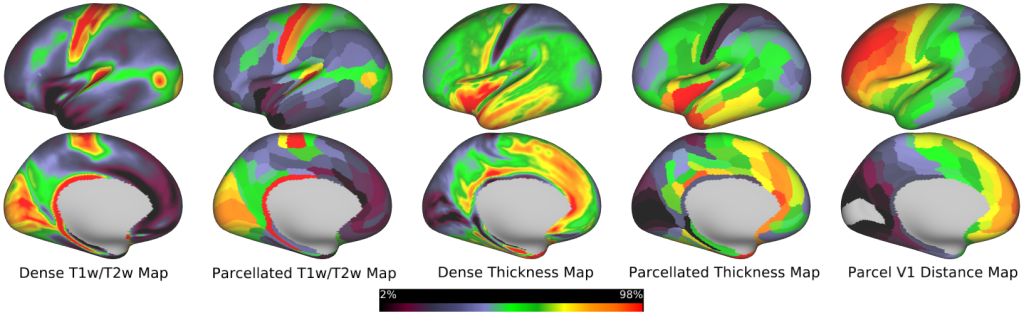
Human Structural Neuroimaging Maps -- Dense and parcellated views of the human neuroimaging maps (T1w/T2w, cortical thickness, and V1 distance maps) used in the study (Figs. 1a, 5a, 6a,d; Supplementary Fig. 1a).
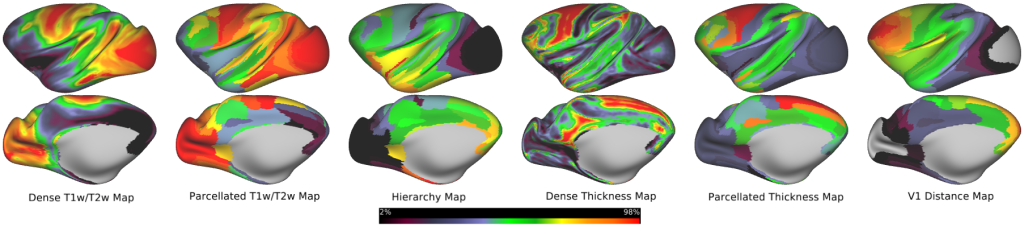
Monkey Structural Neuroimaging Maps -- Dense and parcellated views of the monkey neuroimaging maps (T1w/T2w, cortical thickness, and V1 distance maps) used in the study (Figs. 1c,e, 2a,b; Supplementary Fig. 1b).
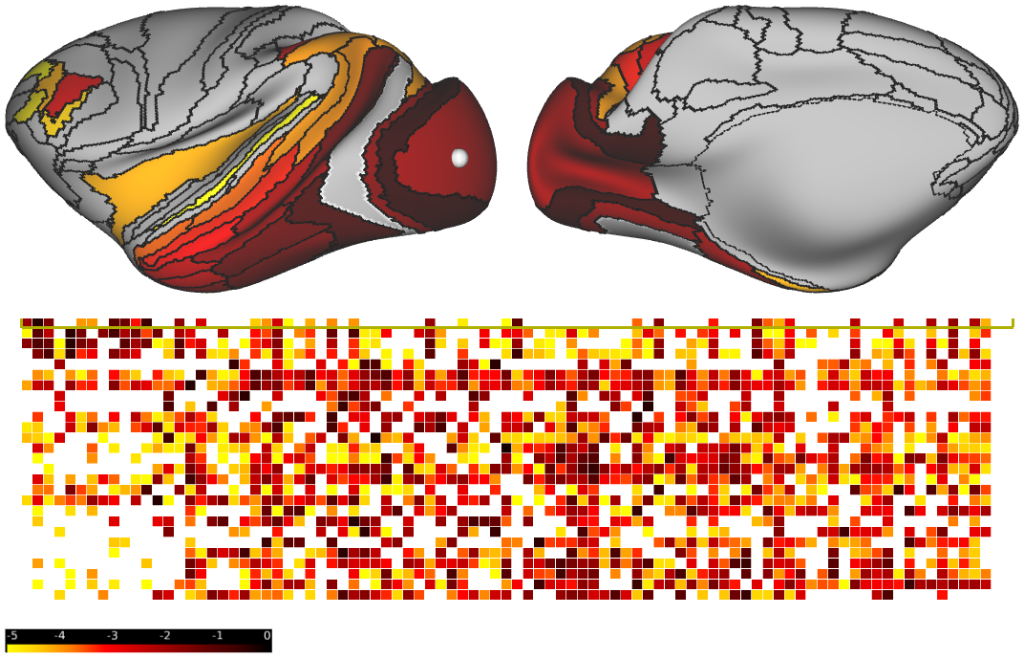
Monkey Interareal Measures -- Interareal neuroanatomical tract-tracing measures of long-range projection strength and laminar specificity (Supplementary Fig. 1a,b), and a non-invasive neuroimaging-derived measure of structural dissimilarity (Fig. 1d) in monkey cortex.
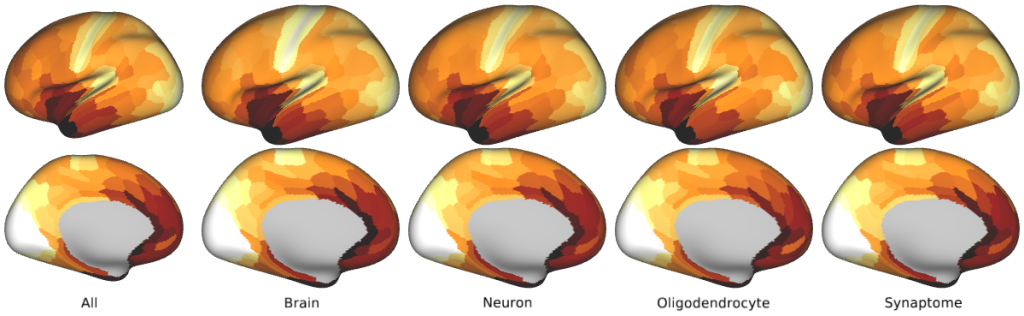
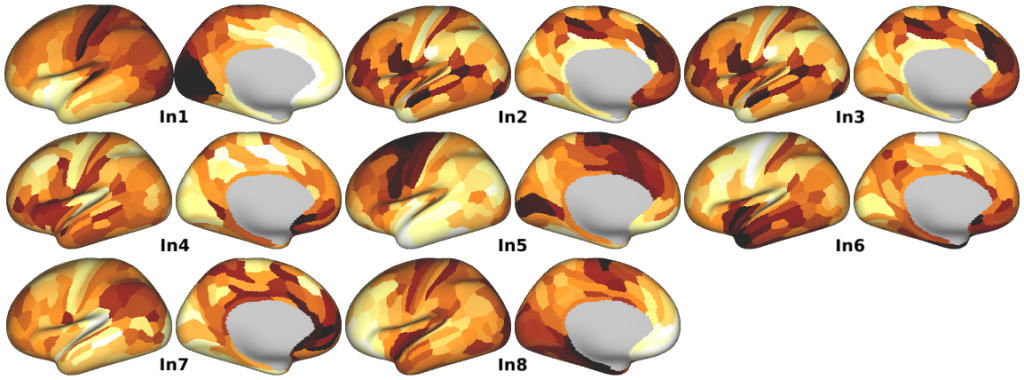
Human Gene Set PCA -- The first principal component (PC1) for the five categorical gene sets used in the study (in Figs. 5 and 6, Supplementary Figs. 6, 8, 9a, and 10).
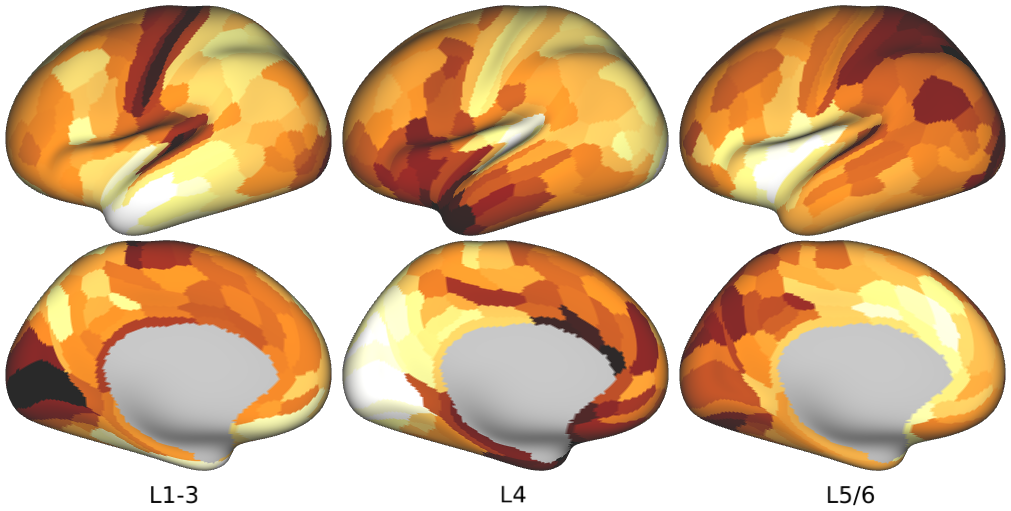
Human Layer-Specific Gene Expression -- Average expression maps of laminar-specific genes in humans (Fig. 4b,c, Supplementary Fig. 10).
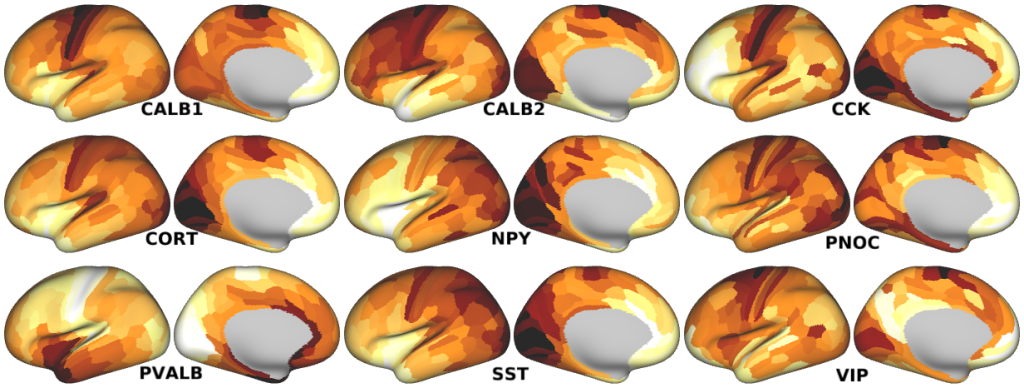
Human Interneuron Marker Gene Expression -- Expression maps of genes coding for proteins associated with different inhibitory interneurons in humans (Fig. 4e,f; Supplementary Fig. 4a).
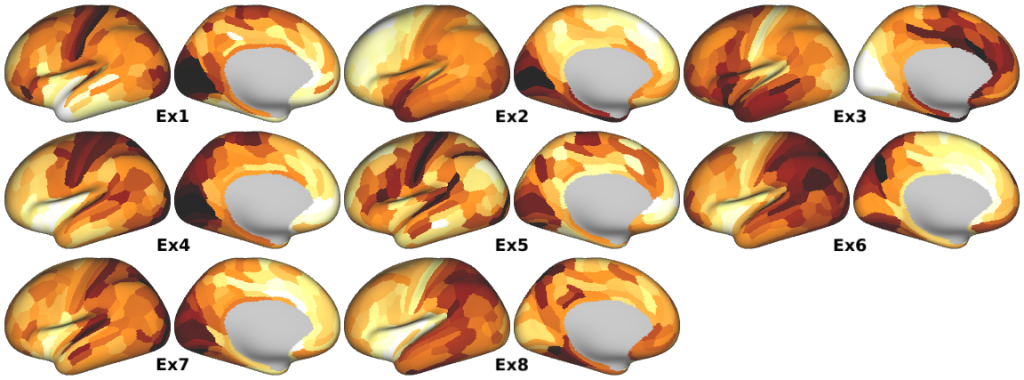
Human Excitatory Cell Type Expression Profiles -- Weighted gene expression profiles characteristic of distinct excitatory neuronal cell types derived from single-cell RNA sequencing of human cortical neurons (Supplementary Fig. 4b).
Human Inhibitory Cell Type Expression Profiles -- Weighted gene expression profiles characteristic of distinct inhibitory neuronal cell types derived from single-cell RNA sequencing of human cortical neurons (Supplementary Fig. 4c).
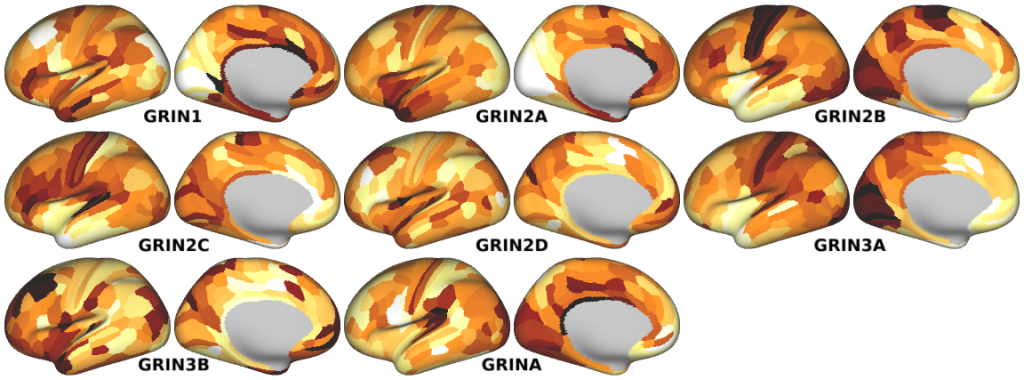
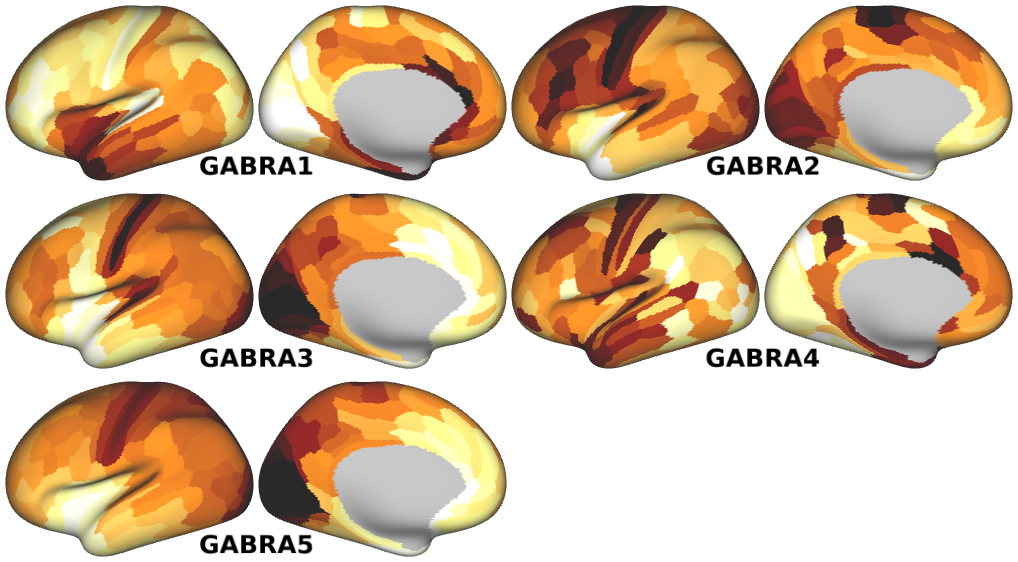
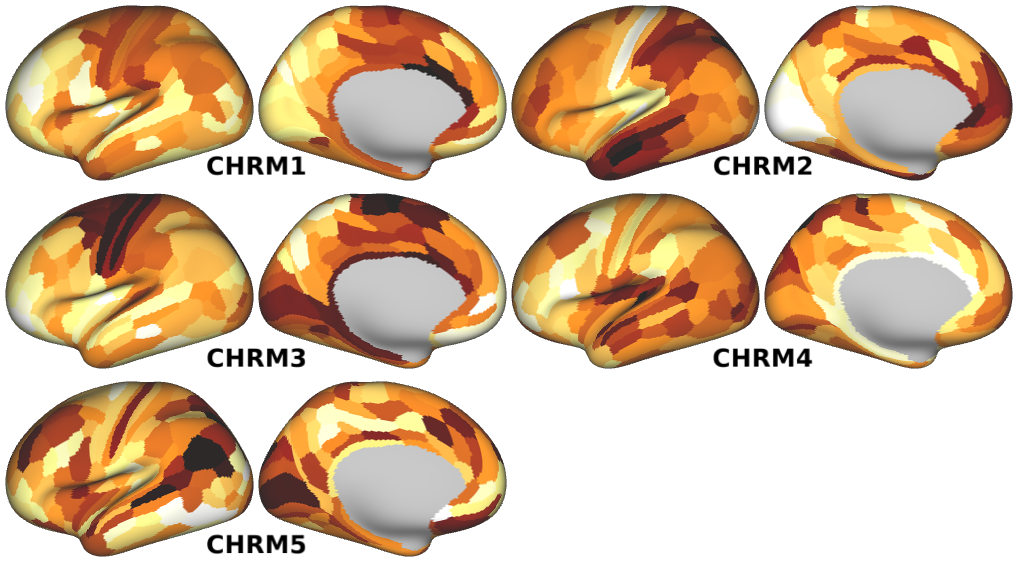
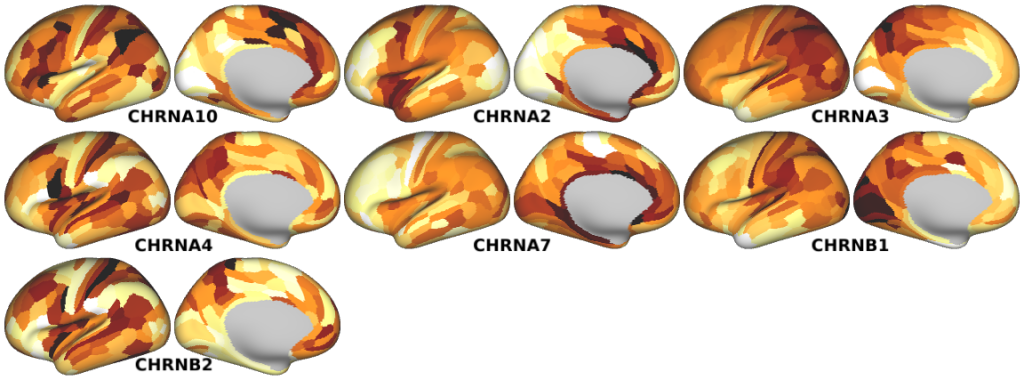
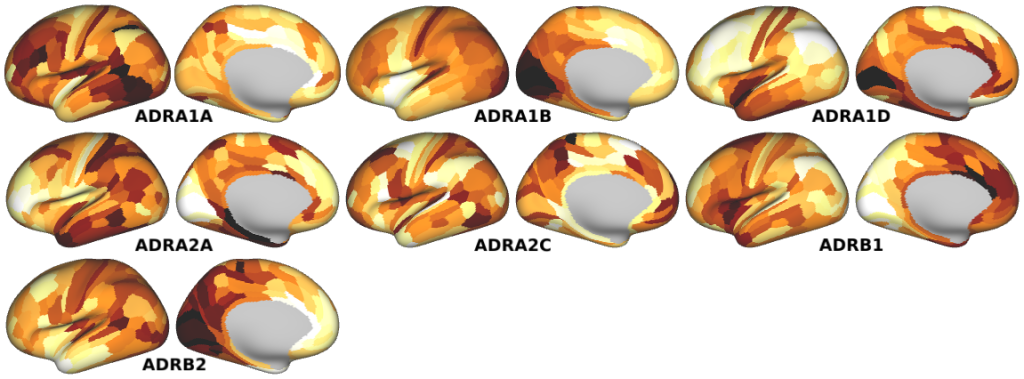
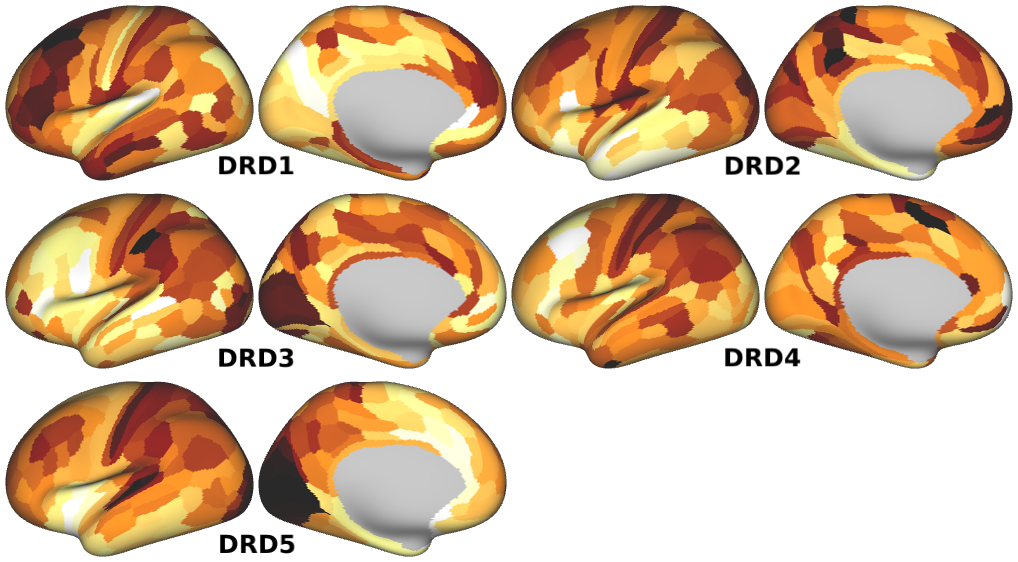
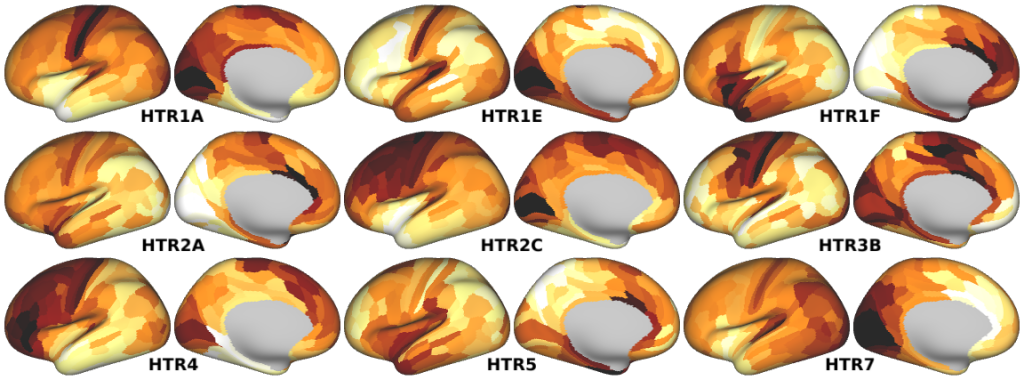
NMDA, GABA_A, Muscarinic and Nicotinic Acetylcholine, Norepinephrine, Dopamine, and Serotonin Receptor Expression Profiles -- Expression profiles for genes coding for various receptor subunits in human cortex (Fig. 4h,i,j; Supplementary Fig. 5).
ABSTRACT:
Hierarchy provides a unifying principle for the macroscale organization of anatomical and functional properties across primate cortex, yet microscale bases of specialization across human cortex are poorly understood. Anatomical hierarchy is conventionally informed by invasive tract-tracing measurements, creating a need for a principled proxy measure in humans. Moreover, cortex exhibits marked interareal variation in gene expression, yet organizing principles of cortical transcription remain unclear. We hypothesized that specialization of cortical microcircuitry involves hierarchical gradients of gene expression. We found that a noninvasive neuroimaging measure-MRI-derived T1-weighted/T2-weighted (T1w/T2w) mapping-reliably indexes anatomical hierarchy, and it captures the dominant pattern of transcriptional variation across human cortex. We found hierarchical gradients in expression profiles of genes related to microcircuit function, consistent with monkey microanatomy, and implicated in neuropsychiatric disorders. Our findings identify a hierarchical axis linking cortical transcription and anatomy, along which gradients of microscale properties may contribute to the macroscale specialization of cortical function.
PUBLICATION:
Nature Neuroscience
- DOI:
10.1038/s41593-018-0195-0
- PMID:
30082915
- Joshua B Burt
- Murat Demirtaş
- William J Eckner
- Natasha M Navejar
- Jie Lisa Ji
- William J Martin
- Alberto Bernacchia
- Alan Anticevic
- John D Murray
- Tulane University
- Yale University
- University of Cambridge
- BlackThorn Therapeutics
-
Burt_et_al_2018_Hierarchy.scene
SCENES:- Cortical Parcellations
- Human Structural Neuroimaging Maps
- Monkey Structural Neuroimaging Maps
- Monkey Interareal Measures
- Human Gene Set PCA
- Human Layer-Specific Gene Expression
- Human Interneuron Marker Gene Expression
- Human Excitatory Cell Type Expression Profiles
- Human Inhibitory Cell Type Expression Profiles
- NMDA Receptor Subunit Expression Profiles
- GABA_A Receptor Subunit Expression Profiles
- Muscarinic Acetylcholine Receptor Expression Profiles
- Nicotinic Acetylcholine Receptor Expression Profiles
- Norepinephrine Receptor Expression Profiles
- Dopamine Receptor Expression Profiles
- Serotonin Receptor Expression Profiles